Molecular virology of hepatitis E virus
- PMID: 21345356
- PMCID: PMC3130092
- DOI: 10.1016/j.virusres.2011.02.011
Molecular virology of hepatitis E virus
Abstract
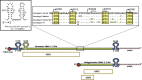
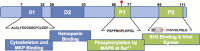
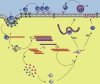
This review details the molecular virology of the hepatitis E virus (HEV). While replicons and in vitro infection systems have recently become available, a lot of information on HEV has been generated through comparisons with better-studied positive-strand RNA viruses and through subgenomic expression of viral open reading frames. These models are now being verified with replicon and infection systems. We provide here the current knowledge on the HEV genome and its constituent proteins--ORF1, ORF2 and ORF3. Based on the available information, we also modify the existing model of the HEV life cycle.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures


References
-
- Agrawal S., Gupta D., Panda S.K. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp) Virology. 2001;282:87–101. - PubMed
-
- Aguiar R.C., Takeyama K., He C., Kreinbrink K., Shipp M.A. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. - PubMed
-
- Ansari I.H., Nanda S.K., Durgapal H., Agrawal S., Mohanty S.K., Gupta D., Jameel S., Panda S.K. Cloning, sequencing, and expression of the hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1) J. Med. Virol. 2000;60:275–283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

