Fold and function of the InlB B-repeat
- PMID: 21345802
- PMCID: PMC3083156
- DOI: 10.1074/jbc.M110.189951
Fold and function of the InlB B-repeat
Abstract
Host cell invasion by the facultative intracellular pathogen Listeria monocytogenes requires the invasion protein InlB in many cell types. InlB consists of an N-terminal internalin domain that binds the host cell receptor tyrosine kinase Met and C-terminal GW domains that bind to glycosaminoglycans (GAGs). Met binding and activation is required for host cell invasion, while the interaction between GW domains and GAGs enhances this effect. Soluble InlB elicits the same cellular phenotypes as the natural Met ligand hepatocyte growth factor/scatter factor (HGF/SF), e.g. cell scatter. So far, little is known about the central part of InlB, the B-repeat. Here we present a structural and functional characterization of the InlB B-repeat. The crystal structure reveals a variation of the β-grasp fold that is most similar to small ubiquitin-like modifiers (SUMOs). However, structural similarity also suggests a potential evolutionary relation to bacterial mucin-binding proteins. The B-repeat defines the prototype structure of a hitherto uncharacterized domain present in over a thousand bacterial proteins. Generally, this domain probably acts as a spacer or a receptor-binding domain in extracellular multi-domain proteins. In cellular assays the B-repeat acts synergistically with the internalin domain conferring to it the ability to stimulate cell motility. Thus, the B-repeat probably binds a further host cell receptor and thereby enhances signaling downstream of Met.
Figures


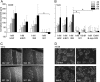



Similar articles
-
MET-activating Residues in the B-repeat of the Listeria monocytogenes Invasion Protein InlB.J Biol Chem. 2016 Dec 2;291(49):25567-25577. doi: 10.1074/jbc.M116.746685. Epub 2016 Oct 27. J Biol Chem. 2016. PMID: 27789707 Free PMC article.
-
Crystal structure of an engineered YopM-InlB hybrid protein.BMC Struct Biol. 2014 Mar 27;14:12. doi: 10.1186/1472-6807-14-12. BMC Struct Biol. 2014. PMID: 24669959 Free PMC article.
-
Structure of the human receptor tyrosine kinase met in complex with the Listeria invasion protein InlB.Cell. 2007 Jul 27;130(2):235-46. doi: 10.1016/j.cell.2007.05.037. Cell. 2007. PMID: 17662939
-
Structural basis of MET receptor dimerization by the bacterial invasion protein InlB and the HGF/SF splice variant NK1.Biochim Biophys Acta. 2013 Oct;1834(10):2195-204. doi: 10.1016/j.bbapap.2012.10.012. Epub 2012 Oct 31. Biochim Biophys Acta. 2013. PMID: 23123275 Review.
-
Structural insights into Met receptor activation.Eur J Cell Biol. 2011 Nov;90(11):972-81. doi: 10.1016/j.ejcb.2010.11.014. Epub 2011 Jan 15. Eur J Cell Biol. 2011. PMID: 21242015 Review.
Cited by
-
Gardnerella vaginalis, Fannyhessea vaginae, and Prevotella bivia Strongly Influence Each Other's Transcriptome in Triple-Species Biofilms.Microb Ecol. 2024 Sep 19;87(1):117. doi: 10.1007/s00248-024-02433-9. Microb Ecol. 2024. PMID: 39294302 Free PMC article.
-
Proteomic Characterization of the Oral Pathogen Filifactor alocis Reveals Key Inter-Protein Interactions of Its RTX Toxin: FtxA.Pathogens. 2022 May 17;11(5):590. doi: 10.3390/pathogens11050590. Pathogens. 2022. PMID: 35631111 Free PMC article.
-
Two Different Species of Mycoplasma Endosymbionts Can Influence Trichomonas vaginalis Pathophysiology.mBio. 2022 Jun 28;13(3):e0091822. doi: 10.1128/mbio.00918-22. Epub 2022 May 24. mBio. 2022. PMID: 35608298 Free PMC article.
-
The Membrane Attack Complex/Perforin Superfamily.J Mol Microbiol Biotechnol. 2017;27(4):252-267. doi: 10.1159/000481286. Epub 2017 Nov 17. J Mol Microbiol Biotechnol. 2017. PMID: 29145176 Free PMC article.
-
Genetic Diversity of Listeria monocytogenes Isolated From Three Commercial Tree Fruit Packinghouses and Evidence of Persistent and Transient Contamination.Front Microbiol. 2022 Jan 10;12:756688. doi: 10.3389/fmicb.2021.756688. eCollection 2021. Front Microbiol. 2022. PMID: 35082763 Free PMC article.
References
-
- Allerberger F., Wagner M. (2010) Clin. Microbiol. Infect. 16, 16–23 - PubMed
-
- Hamon M., Bierne H., Cossart P. (2006) Nat. Rev. Microbiol. 4, 423–434 - PubMed
-
- Cossart P., Toledo-Arana A. (2008) Microbes. Infect. 10, 1041–1050 - PubMed
-
- Mostowy S., Cossart P. (2009) Cell. Host Microbe 5, 510–513 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

