Crystal structure of the C-terminal domain of human DNA primase large subunit: implications for the mechanism of the primase-polymerase α switch
- PMID: 21346410
- PMCID: PMC3100874
- DOI: 10.4161/cc.10.6.15010
Crystal structure of the C-terminal domain of human DNA primase large subunit: implications for the mechanism of the primase-polymerase α switch
Abstract
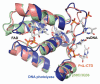
DNA polymerases cannot synthesize DNA without a primer, and DNA primase is the only specialized enzyme capable of de novo synthesis of short RNA primers. In eukaryotes, primase functions within a heterotetrameric complex in concert with a tightly bound DNA polymerase α (Pol α). In humans, the Pol α part is comprised of a catalytic subunit (p180) and an accessory subunit B (p70), and the primase part consists of a small catalytic subunit (p49) and a large essential subunit (p58). The latter subunit participates in primer synthesis, counts the number of nucleotides in a primer, assists the release of the primer-template from primase and transfers it to the Pol α active site. Recently reported crystal structures of the C-terminal domains of the yeast and human enzymes' large subunits provided critical information related to their structure, possible sites for binding of nucleotides and template DNA, as well as the overall organization of eukaryotic primases. However, the structures also revealed a difference in the folding of their proposed DNA-binding fragments, raising the possibility that yeast and human proteins are functionally different. Here we report new structure of the C-terminal domain of the human primase p58 subunit. This structure exhibits a fold similar to a fold reported for the yeast protein but different than a fold reported for the human protein. Based on a comparative analysis of all three C-terminal domain structures, we propose a mechanism of RNA primer length counting and dissociation of the primer-template from primase by a switch in conformation of the ssDNA-binding region of p58.
Figures
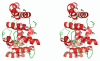

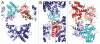

References
-
- Kornberg A, Baker TA. DNA replication. New York: WH Freeman; 1992.
-
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40:115–128. - PubMed
-
- Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. - PubMed
-
- Copeland WC, Wang TS. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem. 1993;268:26179–26189. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
