Analysis of common and specific mechanisms of liver function affected by nitrotoluene compounds
- PMID: 21346803
- PMCID: PMC3035612
- DOI: 10.1371/journal.pone.0014662
Analysis of common and specific mechanisms of liver function affected by nitrotoluene compounds
Abstract
Background: Nitrotoluenes are widely used chemical manufacturing and munitions applications. This group of chemicals has been shown to cause a range of effects from anemia and hypercholesterolemia to testicular atrophy. We have examined the molecular and functional effects of five different, but structurally related, nitrotoluenes on using an integrative systems biology approach to gain insight into common and disparate mechanisms underlying effects caused by these chemicals.
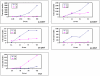
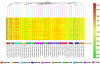
Methodology/principal findings: Sprague-Dawley female rats were exposed via gavage to one of five concentrations of one of five nitrotoluenes [2,4,6-trinitrotoluene (TNT), 2-amino-4,6-dinitrotoluene (2ADNT) 4-amino-2,6-dinitrotoulene (4ADNT), 2,4-dinitrotoluene (2,4DNT) and 2,6-dinitrotoluene (2,6DNT)] with necropsy and tissue collection at 24 or 48 h. Gene expression profile results correlated well with clinical data and liver histopathology that lead to the concept that hematotoxicity was followed by hepatotoxicity. Overall, 2,4DNT, 2,6DNT and TNT had stronger effects than 2ADNT and 4ADNT. Common functional terms, gene expression patterns, pathways and networks were regulated across all nitrotoluenes. These pathways included NRF2-mediated oxidative stress response, aryl hydrocarbon receptor signaling, LPS/IL-1 mediated inhibition of RXR function, xenobiotic metabolism signaling and metabolism of xenobiotics by cytochrome P450. One biological process common to all compounds, lipid metabolism, was found to be impacted both at the transcriptional and lipid production level.
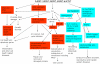
Conclusions/significance: A systems biology strategy was used to identify biochemical pathways affected by five nitroaromatic compounds and to integrate data that tie biochemical alterations to pathological changes. An integrative graphical network model was constructed by combining genomic, gene pathway, lipidomic, and physiological endpoint results to better understand mechanisms of liver toxicity and physiological endpoints affected by these compounds.
Conflict of interest statement
Figures








References
-
- Jenkins TF, Hewitt AD, Grant CL, Thiboutot S, Ampleman G, et al. Identity and distribution of residues of energetic compounds at army live-fire training ranges. Chemosphere. 2006;63:1280–1290. - PubMed
-
- Wintz H, Yoo LJ, Loguinov A, Wu YY, Steevens JA, et al. Gene expression profiles in fathead minnow exposed to 2,4-DNT: correlation with toxicity in mammals. Toxicol Sci. 2006;94:71–82. - PubMed
-
- Gust KA, Pirooznia M, Quinn MJ, Jr, Johnson MS, Escalon L, et al. Neurotoxicogenomic investigations to assess mechanisms of action of the munitions constituents RDX and 2,6-DNT in Northern bobwhite (Colinus virginianus). Toxicol Sci. 2009;110:168–180. - PubMed
-
- Levine BS, Furedi EM, Gordon DE, Barkley JJ, Lish PM. Toxic interactions of the munitions compounds TNT and RDX in F344 rats. Fundam Appl Toxicol. 1990;15:373–380. - PubMed
-
- Talmage SS, Opresko DM, Maxwell CJ, Welsh CJ, Cretella FM, et al. Nitroaromatic munition compounds: environmental effects and screening values. Rev Environ Contam Toxicol. 1999;161:1–156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

