Characterization of the contradictory chromatin signatures at the 3' exons of zinc finger genes
- PMID: 21347206
- PMCID: PMC3039671
- DOI: 10.1371/journal.pone.0017121
Characterization of the contradictory chromatin signatures at the 3' exons of zinc finger genes
Abstract
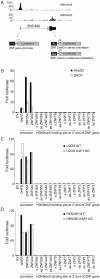
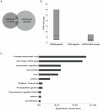
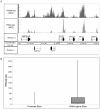
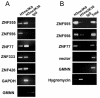
The H3K9me3 histone modification is often found at promoter regions, where it functions to repress transcription. However, we have previously shown that 3' exons of zinc finger genes (ZNFs) are marked by high levels of H3K9me3. We have now further investigated this unusual location for H3K9me3 in ZNF genes. Neither bioinformatic nor experimental approaches support the hypothesis that the 3' exons of ZNFs are promoters. We further characterized the histone modifications at the 3' ZNF exons and found that these regions also contain H3K36me3, a mark of transcriptional elongation. A genome-wide analysis of ChIP-seq data revealed that ZNFs constitute the majority of genes that have high levels of both H3K9me3 and H3K36me3. These results suggested the possibility that the ZNF genes may be imprinted, with one allele transcribed and one allele repressed. To test the hypothesis that the contradictory modifications are due to imprinting, we used a SNP analysis of RNA-seq data to demonstrate that both alleles of certain ZNF genes having H3K9me3 and H3K36me3 are transcribed. We next analyzed isolated ZNF 3' exons using stably integrated episomes. We found that although the H3K36me3 mark was lost when the 3' ZNF exon was removed from its natural genomic location, the isolated ZNF 3' exons retained the H3K9me3 mark. Thus, the H3K9me3 mark at ZNF 3' exons does not impede transcription and it is regulated independently of the H3K36me3 mark. Finally, we demonstrate a strong relationship between the number of tandemly repeated domains in the 3' exons and the H3K9me3 mark. We suggest that the H3K9me3 at ZNF 3' exons may function to protect the genome from inappropriate recombination rather than to regulate transcription.
Conflict of interest statement
Figures



References
-
- Crews D. Epigenetics, brain, behavior, and the environment. Hormones (Athens) 2010;9:41–50. - PubMed
-
- Grayson DR, Chen Y, Dong E, Kundakovic M, Guidotti A. From trans-methylation to cytosine methylation: evolution of the methylation hypothesis of schizophrenia. Epigenetics. 2009;4:144–149. - PubMed
-
- Iwamoto K, Kato T. Epigenetic profiling in schizophrenia and major mental disorders. Neuropsychobiology. 2009;60:5–11. - PubMed

