Improved innate and adaptive immunostimulation by genetically modified HIV-1 protein expressing NYVAC vectors
- PMID: 21347234
- PMCID: PMC3039654
- DOI: 10.1371/journal.pone.0016819
Improved innate and adaptive immunostimulation by genetically modified HIV-1 protein expressing NYVAC vectors
Abstract
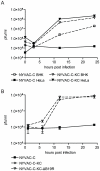
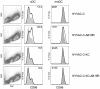
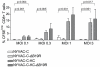
Attenuated poxviruses are safe and capable of expressing foreign antigens. Poxviruses are applied in veterinary vaccination and explored as candidate vaccines for humans. However, poxviruses express multiple genes encoding proteins that interfere with components of the innate and adaptive immune response. This manuscript describes two strategies aimed to improve the immunogenicity of the highly attenuated, host-range restricted poxvirus NYVAC: deletion of the viral gene encoding type-I interferon-binding protein and development of attenuated replication-competent NYVAC. We evaluated these newly generated NYVAC mutants, encoding HIV-1 env, gag, pol and nef, for their ability to stimulate HIV-specific CD8 T-cell responses in vitro from blood mononuclear cells of HIV-infected subjects. The new vectors were evaluated and compared to the parental NYVAC vector in dendritic cells (DCs), RNA expression arrays, HIV gag expression and cross-presentation assays in vitro. Deletion of type-I interferon-binding protein enhanced expression of interferon and interferon-induced genes in DCs, and increased maturation of infected DCs. Restoration of replication competence induced activation of pathways involving antigen processing and presentation. Also, replication-competent NYVAC showed increased Gag expression in infected cells, permitting enhanced cross-presentation to HIV-specific CD8 T cells and proliferation of HIV-specific memory CD8 T-cells in vitro. The recombinant NYVAC combining both modifications induced interferon-induced genes and genes involved in antigen processing and presentation, as well as increased Gag expression. This combined replication-competent NYVAC is a promising candidate for the next generation of HIV vaccines.
Conflict of interest statement
Figures




Similar articles
-
HIV/AIDS Vaccine Candidates Based on Replication-Competent Recombinant Poxvirus NYVAC-C-KC Expressing Trimeric gp140 and Gag-Derived Virus-Like Particles or Lacking the Viral Molecule B19 That Inhibits Type I Interferon Activate Relevant HIV-1-Specific B and T Cell Immune Functions in Nonhuman Primates.J Virol. 2017 Apr 13;91(9):e02182-16. doi: 10.1128/JVI.02182-16. Print 2017 May 1. J Virol. 2017. PMID: 28179536 Free PMC article.
-
Interleukin-1- and type I interferon-dependent enhanced immunogenicity of an NYVAC-HIV-1 Env-Gag-Pol-Nef vaccine vector with dual deletions of type I and type II interferon-binding proteins.J Virol. 2015 Apr;89(7):3819-32. doi: 10.1128/JVI.03061-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609807 Free PMC article.
-
Virological and immunological characterization of novel NYVAC-based HIV/AIDS vaccine candidates expressing clade C trimeric soluble gp140(ZM96) and Gag(ZM96)-Pol-Nef(CN54) as virus-like particles.J Virol. 2015 Jan 15;89(2):970-88. doi: 10.1128/JVI.02469-14. Epub 2014 Oct 29. J Virol. 2015. PMID: 25355891 Free PMC article.
-
Highly attenuated poxvirus vectors: NYVAC, ALVAC and TROVAC.Dev Biol Stand. 1995;84:159-63. Dev Biol Stand. 1995. PMID: 7796949 Review.
-
The interaction of immunodeficiency viruses with dendritic cells.Curr Top Microbiol Immunol. 2003;276:1-30. doi: 10.1007/978-3-662-06508-2_1. Curr Top Microbiol Immunol. 2003. PMID: 12797441 Review.
Cited by
-
Improving Adaptive and Memory Immune Responses of an HIV/AIDS Vaccine Candidate MVA-B by Deletion of Vaccinia Virus Genes (C6L and K7R) Blocking Interferon Signaling Pathways.PLoS One. 2013 Jun 27;8(6):e66894. doi: 10.1371/journal.pone.0066894. Print 2013. PLoS One. 2013. PMID: 23826170 Free PMC article.
-
Removal of vaccinia virus genes that block interferon type I and II pathways improves adaptive and memory responses of the HIV/AIDS vaccine candidate NYVAC-C in mice.J Virol. 2012 May;86(9):5026-38. doi: 10.1128/JVI.06684-11. Epub 2012 Mar 14. J Virol. 2012. PMID: 22419805 Free PMC article.
-
The use of viral vectors in vaccine development.NPJ Vaccines. 2022 Jul 4;7(1):75. doi: 10.1038/s41541-022-00503-y. NPJ Vaccines. 2022. PMID: 35787629 Free PMC article. Review.
-
Vaccinomics, adversomics, and the immune response network theory: individualized vaccinology in the 21st century.Semin Immunol. 2013 Apr;25(2):89-103. doi: 10.1016/j.smim.2013.04.007. Epub 2013 Jun 5. Semin Immunol. 2013. PMID: 23755893 Free PMC article. Review.
-
The evolution of poxvirus vaccines.Viruses. 2015 Apr 7;7(4):1726-803. doi: 10.3390/v7041726. Viruses. 2015. PMID: 25853483 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

