Parallel evolution of a type IV secretion system in radiating lineages of the host-restricted bacterial pathogen Bartonella
- PMID: 21347280
- PMCID: PMC3037411
- DOI: 10.1371/journal.pgen.1001296
Parallel evolution of a type IV secretion system in radiating lineages of the host-restricted bacterial pathogen Bartonella
Abstract
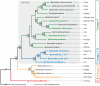
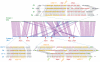
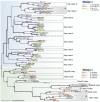
Adaptive radiation is the rapid origination of multiple species from a single ancestor as the result of concurrent adaptation to disparate environments. This fundamental evolutionary process is considered to be responsible for the genesis of a great portion of the diversity of life. Bacteria have evolved enormous biological diversity by exploiting an exceptional range of environments, yet diversification of bacteria via adaptive radiation has been documented in a few cases only and the underlying molecular mechanisms are largely unknown. Here we show a compelling example of adaptive radiation in pathogenic bacteria and reveal their genetic basis. Our evolutionary genomic analyses of the α-proteobacterial genus Bartonella uncover two parallel adaptive radiations within these host-restricted mammalian pathogens. We identify a horizontally-acquired protein secretion system, which has evolved to target specific bacterial effector proteins into host cells as the evolutionary key innovation triggering these parallel adaptive radiations. We show that the functional versatility and adaptive potential of the VirB type IV secretion system (T4SS), and thereby translocated Bartonella effector proteins (Beps), evolved in parallel in the two lineages prior to their radiations. Independent chromosomal fixation of the virB operon and consecutive rounds of lineage-specific bep gene duplications followed by their functional diversification characterize these parallel evolutionary trajectories. Whereas most Beps maintained their ancestral domain constitution, strikingly, a novel type of effector protein emerged convergently in both lineages. This resulted in similar arrays of host cell-targeted effector proteins in the two lineages of Bartonella as the basis of their independent radiation. The parallel molecular evolution of the VirB/Bep system displays a striking example of a key innovation involved in independent adaptive processes and the emergence of bacterial pathogens. Furthermore, our study highlights the remarkable evolvability of T4SSs and their effector proteins, explaining their broad application in bacterial interactions with the environment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

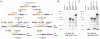
References
-
- Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press; 2000. p. viii, 288.
-
- Grant PR, Grant BR. How and why species multiply : the radiation of Darwin's finches. Princeton: Princeton University Press; 2008. p. xix, 218.
-
- Salzburger W. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol Ecol. 2009;18:169–185. - PubMed
-
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. - PubMed
-
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

