The expansion of the PRAME gene family in Eutheria
- PMID: 21347312
- PMCID: PMC3037382
- DOI: 10.1371/journal.pone.0016867
The expansion of the PRAME gene family in Eutheria
Abstract
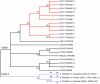
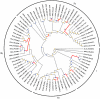
The PRAME gene family belongs to the group of cancer/testis genes whose expression is restricted primarily to the testis and a variety of cancers. The expansion of this gene family as a result of gene duplication has been observed in primates and rodents. We analyzed the PRAME gene family in Eutheria and discovered a novel Y-linked PRAME gene family in bovine, PRAMEY, which underwent amplification after a lineage-specific, autosome-to-Y transposition. Phylogenetic analyses revealed two major evolutionary clades. Clade I containing the amplified PRAMEYs and the unamplified autosomal homologs in cattle and other eutherians is under stronger functional constraints; whereas, Clade II containing the amplified autosomal PRAMEs is under positive selection. Deep-sequencing analysis indicated that eight of the identified 16 PRAMEY loci are active transcriptionally. Compared to the bovine autosomal PRAME that is expressed predominantly in testis, the PRAMEY gene family is expressed exclusively in testis and is up-regulated during testicular maturation. Furthermore, the sense RNA of PRAMEY is expressed specifically whereas the antisense RNA is expressed predominantly in spermatids. This study revealed that the expansion of the PRAME family occurred in both autosomes and sex chromosomes in a lineage-dependent manner. Differential selection forces have shaped the evolution and function of the PRAME family. The positive selection observed on the autosomal PRAMEs (Clade II) may result in their functional diversification in immunity and reproduction. Conversely, selective constraints have operated on the expanded PRAMEYs to preserve their essential function in spermatogenesis.
Conflict of interest statement
Figures



References
-
- Simpson AJG, Caballero OL, Jungbluth A, Chen Y, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer. 2005;5:615–625. - PubMed
-
- Ikeda H, Lethé B, Lehmann F, van Baren N, Baurain JF, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. - PubMed
-
- Peikert T, Specks U, Farver C, Erzurum SC, Comhair SAA. Melanoma antigen A4 is expressed in non-small cell lung cancers and promotes apoptosis. Cancer Res. 2006;66:4693–4700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

