De-novo discovery of differentially abundant transcription factor binding sites including their positional preference
- PMID: 21347314
- PMCID: PMC3037384
- DOI: 10.1371/journal.pcbi.1001070
De-novo discovery of differentially abundant transcription factor binding sites including their positional preference
Erratum in
- PLoS Comput Biol. 2011 Oct;7(10). doi: 10.1371/annotation/a0b541dc-472b-4076-a435-499ce9519335
Abstract
Transcription factors are a main component of gene regulation as they activate or repress gene expression by binding to specific binding sites in promoters. The de-novo discovery of transcription factor binding sites in target regions obtained by wet-lab experiments is a challenging problem in computational biology, which has not been fully solved yet. Here, we present a de-novo motif discovery tool called Dispom for finding differentially abundant transcription factor binding sites that models existing positional preferences of binding sites and adjusts the length of the motif in the learning process. Evaluating Dispom, we find that its prediction performance is superior to existing tools for de-novo motif discovery for 18 benchmark data sets with planted binding sites, and for a metazoan compendium based on experimental data from micro-array, ChIP-chip, ChIP-DSL, and DamID as well as Gene Ontology data. Finally, we apply Dispom to find binding sites differentially abundant in promoters of auxin-responsive genes extracted from Arabidopsis thaliana microarray data, and we find a motif that can be interpreted as a refined auxin responsive element predominately positioned in the 250-bp region upstream of the transcription start site. Using an independent data set of auxin-responsive genes, we find in genome-wide predictions that the refined motif is more specific for auxin-responsive genes than the canonical auxin-responsive element. In general, Dispom can be used to find differentially abundant motifs in sequences of any origin. However, the positional distribution learned by Dispom is especially beneficial if all sequences are aligned to some anchor point like the transcription start site in case of promoter sequences. We demonstrate that the combination of searching for differentially abundant motifs and inferring a position distribution from the data is beneficial for de-novo motif discovery. Hence, we make the tool freely available as a component of the open-source Java framework Jstacs and as a stand-alone application at http://www.jstacs.de/index.php/Dispom.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
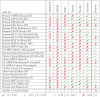
 , and we use the thresholds on the normalized euclidean distance as proposed by Linhart et al. . The
, and we use the thresholds on the normalized euclidean distance as proposed by Linhart et al. . The  symbol marks long execution times (
symbol marks long execution times ( h) that were aborted in . In the last row of the table, we report the total number of motifs discovered by each of the tools.
h) that were aborted in . In the last row of the table, we report the total number of motifs discovered by each of the tools.
 and
and  below 0.1 for all available thresholds. These curves would be located in the lower left corner of both subfigures.
below 0.1 for all available thresholds. These curves would be located in the lower left corner of both subfigures.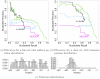


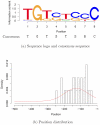
 and
and  .
.
Similar articles
-
Dispom: a discriminative de-novo motif discovery tool based on the jstacs library.J Bioinform Comput Biol. 2013 Feb;11(1):1340006. doi: 10.1142/S0219720013400064. Epub 2013 Jan 21. J Bioinform Comput Biol. 2013. PMID: 23427988
-
Positional distribution of transcription factor binding sites in Arabidopsis thaliana.Sci Rep. 2016 Apr 27;6:25164. doi: 10.1038/srep25164. Sci Rep. 2016. PMID: 27117388 Free PMC article.
-
Sequence-based prediction of transcription upregulation by auxin in plants.J Bioinform Comput Biol. 2015 Feb;13(1):1540009. doi: 10.1142/S0219720015400090. J Bioinform Comput Biol. 2015. PMID: 25666655
-
From experiment-driven database analyses to database-driven experiments in Arabidopsis thaliana transcription factor research.Plant Sci. 2017 Sep;262:141-147. doi: 10.1016/j.plantsci.2017.06.011. Epub 2017 Jun 27. Plant Sci. 2017. PMID: 28716409 Review.
-
Computational framework for the prediction of transcription factor binding sites by multiple data integration.BMC Neurosci. 2006 Oct 30;7 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2202-7-S1-S8. BMC Neurosci. 2006. PMID: 17118162 Free PMC article. Review.
Cited by
-
Evaluation of methods for modeling transcription factor sequence specificity.Nat Biotechnol. 2013 Feb;31(2):126-34. doi: 10.1038/nbt.2486. Epub 2013 Jan 27. Nat Biotechnol. 2013. PMID: 23354101 Free PMC article.
-
Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome.BMC Genomics. 2014;15 Suppl 12(Suppl 12):S4. doi: 10.1186/1471-2164-15-S12-S4. Epub 2014 Dec 19. BMC Genomics. 2014. PMID: 25563792 Free PMC article.
-
Genomic background sequences systematically outperform synthetic ones in de novo motif discovery for ChIP-seq data.NAR Genom Bioinform. 2024 Jul 27;6(3):lqae090. doi: 10.1093/nargab/lqae090. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39071850 Free PMC article.
-
Binding site discovery from nucleic acid sequences by discriminative learning of hidden Markov models.Nucleic Acids Res. 2014 Dec 1;42(21):12995-3011. doi: 10.1093/nar/gku1083. Epub 2014 Nov 11. Nucleic Acids Res. 2014. PMID: 25389269 Free PMC article.
-
POWRS: position-sensitive motif discovery.PLoS One. 2012;7(7):e40373. doi: 10.1371/journal.pone.0040373. Epub 2012 Jul 5. PLoS One. 2012. PMID: 22792292 Free PMC article.
References
-
- Benotmane AM, Hoylaerts MF, Collen D, Belayew A. Nonisotopic quantitative analysis of protein-DNA interactions at equilibrium. Anal Biochem. 1997;250:181–185. - PubMed
-
- Mönke G, Altschmied L, Tewes A, Reidt W, Mock HP, et al. Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta. 2004;219:158–166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

