Epstein-Barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1
- PMID: 21347341
- PMCID: PMC3037348
- DOI: 10.1371/journal.ppat.1001275
Epstein-Barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1
Abstract
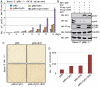
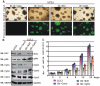
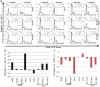
EBNA3C, one of the Epstein-Barr virus (EBV)-encoded latent antigens, is essential for primary B-cell transformation. Cyclin D1, a key regulator of G1 to S phase progression, is tightly associated and aberrantly expressed in numerous human cancers. Previously, EBNA3C was shown to bind to Cyclin D1 in vitro along with Cyclin A and Cyclin E. In the present study, we provide evidence which demonstrates that EBNA3C forms a complex with Cyclin D1 in human cells. Detailed mapping experiments show that a small N-terminal region which lies between amino acids 130-160 of EBNA3C binds to two different sites of Cyclin D1- the N-terminal pRb binding domain (residues 1-50), and C-terminal domain (residues 171-240), known to regulate Cyclin D1 stability. Cyclin D1 is short-lived and ubiquitin-mediated proteasomal degradation has been targeted as a means of therapeutic intervention. Here, we show that EBNA3C stabilizes Cyclin D1 through inhibition of its poly-ubiquitination, and also increases its nuclear localization by blocking GSK3β activity. We further show that EBNA3C enhances the kinase activity of Cyclin D1/CDK6 which enables subsequent ubiquitination and degradation of pRb. EBNA3C together with Cyclin D1-CDK6 complex also efficiently nullifies the inhibitory effect of pRb on cell growth. Moreover, an sh-RNA based strategy for knock-down of both cyclin D1 and EBNA3C genes in EBV transformed lymphoblastoid cell lines (LCLs) shows a significant reduction in cell-growth. Based on these results, we propose that EBNA3C can stabilize as well as enhance the functional activity of Cyclin D1 thereby facilitating the G1-S transition in EBV transformed lymphoblastoid cell lines.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








Similar articles
-
Epstein-Barr Virus Nuclear Antigen 3C Facilitates Cell Proliferation by Regulating Cyclin D2.J Virol. 2018 Aug 29;92(18):e00663-18. doi: 10.1128/JVI.00663-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29997218 Free PMC article.
-
Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2.J Virol. 2009 May;83(9):4652-69. doi: 10.1128/JVI.02408-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244339 Free PMC article.
-
A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein-Barr virus-infected cells.J Virol. 2004 Dec;78(23):12857-67. doi: 10.1128/JVI.78.23.12857-12867.2004. J Virol. 2004. PMID: 15542638 Free PMC article.
-
Nucleoside diphosphate kinase/Nm23 and Epstein-Barr virus.Mol Cell Biochem. 2009 Sep;329(1-2):131-9. doi: 10.1007/s11010-009-0123-4. Epub 2009 May 3. Mol Cell Biochem. 2009. PMID: 19412732 Free PMC article. Review.
-
The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration.Front Biosci. 2002 Mar 1;7:d704-16. doi: 10.2741/subraman. Front Biosci. 2002. PMID: 11861219 Review.
Cited by
-
Screening and functional analysis of differentially expressed genes in EBV-transformed lymphoblasts.Virol J. 2012 Mar 30;9:77. doi: 10.1186/1743-422X-9-77. Virol J. 2012. PMID: 22458412 Free PMC article.
-
Epstein-Barr Virus: Diseases Linked to Infection and Transformation.Front Microbiol. 2016 Oct 25;7:1602. doi: 10.3389/fmicb.2016.01602. eCollection 2016. Front Microbiol. 2016. PMID: 27826287 Free PMC article. Review.
-
An essential EBV latent antigen 3C binds Bcl6 for targeted degradation and cell proliferation.PLoS Pathog. 2017 Jul 24;13(7):e1006500. doi: 10.1371/journal.ppat.1006500. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28738086 Free PMC article.
-
The EBNA3 family of Epstein-Barr virus nuclear proteins associates with the USP46/USP12 deubiquitination complexes to regulate lymphoblastoid cell line growth.PLoS Pathog. 2015 Apr 9;11(4):e1004822. doi: 10.1371/journal.ppat.1004822. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25855980 Free PMC article.
-
NVP-BEZ235, a novel dual PI3K/mTOR inhibitor, enhances the radiosensitivity of human glioma stem cells in vitro.Acta Pharmacol Sin. 2013 May;34(5):681-90. doi: 10.1038/aps.2013.22. Epub 2013 Apr 22. Acta Pharmacol Sin. 2013. PMID: 23603977 Free PMC article.
References
-
- Robertson ES. Norwich, United Kingdom: Caister Academic Press; 2010. Epstein-Barr virus: latency and transformation.
-
- Kieff E, Rickinson AB. Epstein-Barr virus and its replication. In: Knipe D, Howley P, editors. Fields virology, 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 2511–2573.
-
- O'Nions J, Allday MJ. Deregulation of the cell cycle by the Epstein-Barr virus. Adv Cancer Res. 2004;92:119–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

