Functional analysis of an acid adaptive DNA adenine methyltransferase from Helicobacter pylori 26695
- PMID: 21347417
- PMCID: PMC3036652
- DOI: 10.1371/journal.pone.0016810
Functional analysis of an acid adaptive DNA adenine methyltransferase from Helicobacter pylori 26695
Abstract
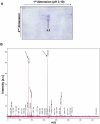
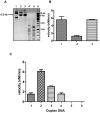
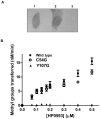
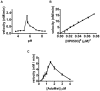
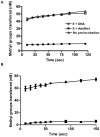
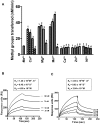
HP0593 DNA-(N(6)-adenine)-methyltransferase (HP0593 MTase) is a member of a Type III restriction-modification system in Helicobacter pylori strain 26695. HP0593 MTase has been cloned, overexpressed and purified heterologously in Escherichia coli. The recognition sequence of the purified MTase was determined as 5'-GCAG-3'and the site of methylation was found to be adenine. The activity of HP0593 MTase was found to be optimal at pH 5.5. This is a unique property in context of natural adaptation of H. pylori in its acidic niche. Dot-blot assay using antibodies that react specifically with DNA containing m6A modification confirmed that HP0593 MTase is an adenine-specific MTase. HP0593 MTase occurred as both monomer and dimer in solution as determined by gel-filtration chromatography and chemical-crosslinking studies. The nonlinear dependence of methylation activity on enzyme concentration indicated that more than one molecule of enzyme was required for its activity. Analysis of initial velocity with AdoMet as a substrate showed that two molecules of AdoMet bind to HP0593 MTase, which is the first example in case of Type III MTases. Interestingly, metal ion cofactors such as Co(2+), Mn(2+), and also Mg(2+) stimulated the HP0593 MTase activity. Preincubation and isotope partitioning analyses clearly indicated that HP0593 MTase-DNA complex is catalytically competent, and suggested that DNA binds to the MTase first followed by AdoMet. HP0593 MTase shows a distributive mechanism of methylation on DNA having more than one recognition site. Considering the occurrence of GCAG sequence in the potential promoter regions of physiologically important genes in H. pylori, our results provide impetus for exploring the role of this DNA MTase in the cellular processes of H. pylori.
Conflict of interest statement
Figures

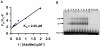
Similar articles
-
N6-Adenosine DNA Methyltransferase from H. pylori 98-10 Strain in Complex with DNA and AdoMet: Structural Insights from in Silico Studies.J Phys Chem B. 2017 Jan 19;121(2):365-378. doi: 10.1021/acs.jpcb.6b08433. Epub 2017 Jan 5. J Phys Chem B. 2017. PMID: 28054779
-
Kinetic and catalytic properties of dimeric KpnI DNA methyltransferase.J Biol Chem. 2003 Mar 7;278(10):7863-74. doi: 10.1074/jbc.M211458200. Epub 2002 Dec 28. J Biol Chem. 2003. PMID: 12506109
-
Biochemical and structural characterization of a DNA N6-adenine methyltransferase from Helicobacter pylori.Oncotarget. 2016 Jul 5;7(27):40965-40977. doi: 10.18632/oncotarget.9692. Oncotarget. 2016. PMID: 27259995 Free PMC article.
-
The Helicobacter pylori Methylome: Roles in Gene Regulation and Virulence.Curr Top Microbiol Immunol. 2017;400:105-127. doi: 10.1007/978-3-319-50520-6_5. Curr Top Microbiol Immunol. 2017. PMID: 28124151 Review.
-
DNA methyltransferases: mechanistic models derived from kinetic analysis.Crit Rev Biochem Mol Biol. 2012 Mar-Apr;47(2):97-193. doi: 10.3109/10409238.2011.620942. Epub 2012 Jan 20. Crit Rev Biochem Mol Biol. 2012. PMID: 22260147 Review.
Cited by
-
Networking and Specificity-Changing DNA Methyltransferases in Helicobacter pylori.Front Microbiol. 2020 Jul 17;11:1628. doi: 10.3389/fmicb.2020.01628. eCollection 2020. Front Microbiol. 2020. PMID: 32765461 Free PMC article.
-
Beta class amino methyltransferases from bacteria to humans: evolution and structural consequences.Nucleic Acids Res. 2020 Oct 9;48(18):10034-10044. doi: 10.1093/nar/gkaa446. Nucleic Acids Res. 2020. PMID: 32453412 Free PMC article. Review.
-
Diverse functions of restriction-modification systems in addition to cellular defense.Microbiol Mol Biol Rev. 2013 Mar;77(1):53-72. doi: 10.1128/MMBR.00044-12. Microbiol Mol Biol Rev. 2013. PMID: 23471617 Free PMC article. Review.
-
Determination of cell uptake pathways for tumor inhibitor lysyl oxidase propeptide.Mol Oncol. 2016 Jan;10(1):1-23. doi: 10.1016/j.molonc.2015.07.005. Epub 2015 Aug 6. Mol Oncol. 2016. PMID: 26297052 Free PMC article.
-
Theoretical study on the binding mechanism between N6-methyladenine and natural DNA bases.J Mol Model. 2013 Mar;19(3):1089-98. doi: 10.1007/s00894-012-1628-4. Epub 2012 Nov 9. J Mol Model. 2013. PMID: 23138643
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

