Specific age-associated DNA methylation changes in human dermal fibroblasts
- PMID: 21347436
- PMCID: PMC3035656
- DOI: 10.1371/journal.pone.0016679
Specific age-associated DNA methylation changes in human dermal fibroblasts
Abstract
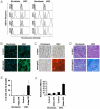
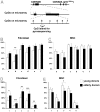
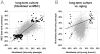
Epigenetic modifications of cytosine residues in the DNA play a critical role for cellular differentiation and potentially also for aging. In mesenchymal stromal cells (MSC) from human bone marrow we have previously demonstrated age-associated methylation changes at specific CpG-sites of developmental genes. In continuation of this work, we have now isolated human dermal fibroblasts from young (<23 years) and elderly donors (>60 years) for comparison of their DNA methylation profiles using the Infinium HumanMethylation27 assay. In contrast to MSC, fibroblasts could not be induced towards adipogenic, osteogenic and chondrogenic lineage and this is reflected by highly significant differences between the two cell types: 766 CpG sites were hyper-methylated and 752 CpG sites were hypo-methylated in fibroblasts in comparison to MSC. Strikingly, global DNA methylation profiles of fibroblasts from the same dermal region clustered closely together indicating that fibroblasts maintain positional memory even after in vitro culture. 75 CpG sites were more than 15% differentially methylated in fibroblasts upon aging. Very high hyper-methylation was observed in the aged group within the INK4A/ARF/INK4b locus and this was validated by pyrosequencing. Age-associated DNA methylation changes were related in fibroblasts and MSC but they were often regulated in opposite directions between the two cell types. In contrast, long-term culture associated changes were very consistent in fibroblasts and MSC. Epigenetic modifications at specific CpG sites support the notion that aging represents a coordinated developmental mechanism that seems to be regulated in a cell type specific manner.
Conflict of interest statement
Figures

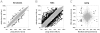
References
-
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. - PubMed
-
- Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007;1100:60–74. - PubMed
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. - PubMed
-
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

