Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis
- PMID: 21347703
- PMCID: PMC3932693
- DOI: 10.1007/s10059-011-0031-9
Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis
Abstract
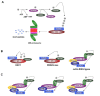
Ubiquitination is a unique protein degradation system utilized by eukaryotes to efficiently degrade detrimental cellular proteins and control the entire pool of regulatory components. In plants, adaptation in response to various abiotic stresses can be achieved through ubiquitination and the resulting degradation of components specific to these stress signalings. Arabidopsis has more than 1,400 E3 enzymes, indicating E3 ligase acts as a main determinant of substrate specificity. However, as only a minority of E3 ligases related to abiotic stress signaling have been studied in Arabidopsis, the further elucidation of the biological roles and related substrates of newly identified E3 ligases is essential in order to clarify the functional relationship between abiotic stress and E3 ligases. Here, we review the current knowledge and future prospects of the regulatory mechanism and role of several E3 ligases involved in abiotic stress signal transduction in Arabidopsis. As another potential approach to understand how ubiquitination is involved in such signaling, we also briefly introduce factors that regulate the activity of cullin in multisubunit E3 ligase complexes.
Figures

References
-
- Agarwal P.K., Agarwal P., Reddy M.K., Sopory S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. (2006);25:1263–1274. - PubMed
-
- Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. (2006);443:590–593. - PubMed
-
- Aravind L., Koonin E.V. The U box is a modified RING finger: a common domain in ubiquitination. Curr. Biol. (2000);10:R132–R134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

