Regulation of mouse 4-1BB expression: multiple promoter usages and a splice variant
- PMID: 21347708
- PMCID: PMC3932682
- DOI: 10.1007/s10059-011-0018-6
Regulation of mouse 4-1BB expression: multiple promoter usages and a splice variant
Abstract
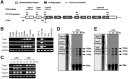
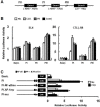
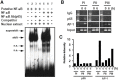
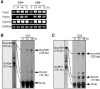
The expression of 4-1BB has been known to be dependent on T cell activation. Recent studies have, however, revealed that 4-1BB expression is not restricted to T cells. We sought to determine the molecular basis for the differential gene expression. Here we report the expression pattern of two mouse 4-1BB transcripts, type I and type II. Whereas the type I transcript was specifically expressed on immune organ as previously reported, the type II transcript was ubiquitously expressed in tissues and various cell lines. However, both type I and type II transcript were highly induced on activated T cells. Primer extension assay of the two 4-1BB transcripts suggested that mouse 4-1BB had more than two transcripts. Using luciferase assay we have identified three promoter regions (PI, PII and PIII), which located on upstream region of second exon 1, first exon 1, and exon 2, respectively. In particular, the type I transcript was preferentially induced when naïve T cells are stimulated by anti-CD3 monoclonal antibody (mAb) since NF-κB specifically binds to the putative NF-κB element of PI. We have also shown that a splice variant, in which the transmembrane domain was deleted, could inhibit 4-1BB signaling. The splicing variant was highly induced by TCR stimulation. Our results reveal 4-1BB also has a negative regulation system through soluble 4-1BB produced from a splice variant induced under activation conditions.
Figures


Similar articles
-
NF-kappaB and AP-1 regulate activation-dependent CD137 (4-1BB) expression in T cells.FEBS Lett. 2003 Apr 24;541(1-3):163-70. doi: 10.1016/s0014-5793(03)00326-0. FEBS Lett. 2003. PMID: 12706838
-
Cross-talk between 4-1BB and TLR1-TLR2 Signaling in CD8+ T Cells Regulates TLR2's Costimulatory Effects.Cancer Immunol Res. 2016 Aug;4(8):708-16. doi: 10.1158/2326-6066.CIR-15-0173. Epub 2016 Jun 7. Cancer Immunol Res. 2016. PMID: 27267778 Free PMC article.
-
4-1BB signaling activates the t cell factor 1 effector/β-catenin pathway with delayed kinetics via ERK signaling and delayed PI3K/AKT activation to promote the proliferation of CD8+ T Cells.PLoS One. 2013 Jul 11;8(7):e69677. doi: 10.1371/journal.pone.0069677. Print 2013. PLoS One. 2013. PMID: 23874982 Free PMC article.
-
A mRNA variant encoding a soluble form of 4-1BB, a member of the murine NGF/TNF receptor family.Gene. 1995 Oct 27;164(2):311-5. doi: 10.1016/0378-1119(95)00349-b. Gene. 1995. PMID: 7590349
-
Biochemical and immunological characteristics of 4-1BB (CD137) receptor and ligand and potential applications in cancer therapy.Arch Immunol Ther Exp (Warsz). 1999;47(5):275-9. Arch Immunol Ther Exp (Warsz). 1999. PMID: 10604232 Review.
Cited by
-
AP-1 Transcription Factors as Regulators of Immune Responses in Cancer.Cancers (Basel). 2019 Jul 23;11(7):1037. doi: 10.3390/cancers11071037. Cancers (Basel). 2019. PMID: 31340499 Free PMC article. Review.
-
The Murine CD137/CD137 Ligand Signalosome: A Signal Platform Generating Signal Complexity.Front Immunol. 2020 Dec 10;11:553715. doi: 10.3389/fimmu.2020.553715. eCollection 2020. Front Immunol. 2020. PMID: 33362756 Free PMC article. Review.
-
Regulation of CD137 expression through K-Ras signaling in pancreatic cancer cells.Cancer Commun (Lond). 2019 Jul 9;39(1):41. doi: 10.1186/s40880-019-0386-4. Cancer Commun (Lond). 2019. PMID: 31288851 Free PMC article.
-
4-1BB signaling breaks the tolerance of maternal CD8+ T cells that are reactive with alloantigens.PLoS One. 2012;7(9):e45481. doi: 10.1371/journal.pone.0045481. Epub 2012 Sep 21. PLoS One. 2012. PMID: 23029041 Free PMC article.
-
Cellular dissection of psoriasis for transcriptome analyses and the post-GWAS era.BMC Med Genomics. 2014 May 22;7:27. doi: 10.1186/1755-8794-7-27. BMC Med Genomics. 2014. PMID: 24885462 Free PMC article.
References
-
- Alderson M.R., Smith C.A., Tough T.W., Davis-Smith T., Armitage R.J., Falk B., Roux E., Baker E., Sutherland G.R., Din W.S. Molecular and biological characterization of human 4-1BB and its ligand. Eur. J. Immunol. (1994);24:2219–2227. - PubMed
-
- Boussaud V., Soler P., Moreau J., Goodwin R.G., Hance A.J. Expression of three members of the TNF-R family of receptors (4-1BB, lymphotoxin-beta receptor, and Fas) in human lung. Eur. Respir. J. (1998);12:926–931. - PubMed
-
- Broll K., Richter G., Pauly S., Hofstaedter F., Schwarz H. CD137 expression in tumor vessel walls. High correlation with malignant tumors. Am. J. Clin. Pathol. (2001);115:543–549. - PubMed
-
- Cascino I., Fiucci G., Papoff G., Ruberti G. Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J. Immunol. (1995);154:2706–2713. - PubMed
-
- Cascino I., Papoff G., De Maria R., Testi R., Ruberti G. Fas/Apo-1 (CD95) receptor lacking the intracytoplasmic signaling domain protects tumor cells from Fas-mediated apoptosis. J. Immunol. (1996);156:13–17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

