Autophagy in immunity and cell-autonomous defense against intracellular microbes
- PMID: 21349088
- PMCID: PMC3057454
- DOI: 10.1111/j.1600-065X.2010.00995.x
Autophagy in immunity and cell-autonomous defense against intracellular microbes
Abstract
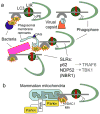
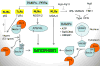
Autophagy was viewed until very recently primarily as a metabolic and intracellular biomass and organelle quality and quantity control pathway. It has now been recognized that autophagy represents a bona fide immunologic process with a wide array of roles in immunity. The immunologic functions of autophagy, as we understand them now, span both innate and adaptive immunity. They range from unique and sometimes highly specialized immunologic effectors and regulatory functions (referred to here as type I immunophagy) to generic homeostatic influence on immune cells (type II immunophagy), akin to the effects on survival and homeostasis of other cell types in the body. As a concept-building tool for understanding why and how autophagy is intertwined with immunity, it is useful to consider that the presently complex picture has emerged in increments, starting in part from the realization that autophagy acts as an evolutionarily ancient microbial clearance mechanism defending eukaryotic cells against intracellular pathogens. In this review, we build a stepwise model of how the core axis of autophagy as a cell-autonomous immune defense against microbes evolved into a complex but orderly web of intersections with innate and adaptive immunity processes. The connections between autophagy and conventional immunity systems include Toll-like receptors, Nod-like receptors, RIG-I-like receptors, damage-associated molecular patterns such as HMGB1, other known innate and adaptive immunity receptors and cytokines, sequestasome (p62)-like receptors that act as autophagy adapters, immunity-related GTPase IRGM, innate and adaptive functions of macrophages and dendritic cells, and differential effects on development and homeostasis of T- and B-lymphocyte subsets. The disease contexts covered here include tuberculosis, infections with human immunodeficiency virus and other viruses, Salmonella, Listeria, Shigella, Toxoplasma, and inflammatory disorders such as Crohn's disease and multiple sclerosis.
© 2011 John Wiley & Sons A/S.
Figures





References
-
- Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–528. - PubMed
-
- Munz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–449. - PubMed
-
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

