The properties and applications of single-molecule DNA sequencing
- PMID: 21349208
- PMCID: PMC3188791
- DOI: 10.1186/gb-2011-12-2-217
The properties and applications of single-molecule DNA sequencing
Abstract
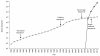
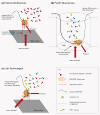
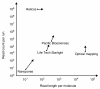
Single-molecule sequencing enables DNA or RNA to be sequenced directly from biological samples, making it well-suited for diagnostic and clinical applications. Here we review the properties and applications of this rapidly evolving and promising technology.
Figures
Similar articles
-
Next generation sequencing: the technology we need in pediatric laboratories?Clin Biochem. 2011 May;44(7):514-515. doi: 10.1016/j.clinbiochem.2011.02.030. Clin Biochem. 2011. PMID: 22036354 No abstract available.
-
Next-generation sequencing and potential applications in fungal genomics.Methods Mol Biol. 2011;722:51-60. doi: 10.1007/978-1-61779-040-9_4. Methods Mol Biol. 2011. PMID: 21590412
-
WHEN TWO IS BETTER THAN ONE.Biotechniques. 2016 Feb 1;60(2):56-60. doi: 10.2144/000114376. eCollection 2016 Feb. Biotechniques. 2016. PMID: 26842349
-
Analyzing Modern Biomolecules: The Revolution of Nucleic-Acid Sequencing - Review.Biomolecules. 2021 Jul 28;11(8):1111. doi: 10.3390/biom11081111. Biomolecules. 2021. PMID: 34439777 Free PMC article. Review.
-
Primer design and primer-directed sequencing.Methods Mol Biol. 2001;167:39-51. doi: 10.1385/1-59259-113-2:039. Methods Mol Biol. 2001. PMID: 11265320 Review. No abstract available.
Cited by
-
Cancer genome sequencing and its implications for personalized cancer vaccines.Cancers (Basel). 2011 Nov 25;3(4):4191-211. doi: 10.3390/cancers3044191. Cancers (Basel). 2011. PMID: 24213133 Free PMC article.
-
SMRT sequencing of full-length transcriptome of seagrasses Zostera japonica.Sci Rep. 2019 Oct 10;9(1):14537. doi: 10.1038/s41598-019-51176-y. Sci Rep. 2019. PMID: 31601990 Free PMC article.
-
Functional Characterization of TNFα in the Starry Flounder (Platichthys stellatus) and Its Potential as an Immunostimulant.Animals (Basel). 2025 Jul 17;15(14):2119. doi: 10.3390/ani15142119. Animals (Basel). 2025. PMID: 40723580 Free PMC article.
-
Identification and Analysis of Candidate Genes Associated with Yield Structure Traits and Maize Yield Using Next-Generation Sequencing Technology.Genes (Basel). 2023 Dec 29;15(1):56. doi: 10.3390/genes15010056. Genes (Basel). 2023. PMID: 38254946 Free PMC article.
-
Modeling of shotgun sequencing of DNA plasmids using experimental and theoretical approaches.BMC Bioinformatics. 2020 Apr 3;21(1):132. doi: 10.1186/s12859-020-3461-6. BMC Bioinformatics. 2020. PMID: 32245400 Free PMC article.
References
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J. et al.Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

