Structure and mechanism of glutamate receptor ion channel assembly, activation and modulation
- PMID: 21349697
- PMCID: PMC3092858
- DOI: 10.1016/j.conb.2011.02.001
Structure and mechanism of glutamate receptor ion channel assembly, activation and modulation
Abstract
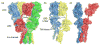
Ionotropic glutamate receptors (iGluRs) are ligand gated ion channels that mediate excitatory synaptic transmission in the brain of vertebrates. A rapidly growing body of crystal structures for isolated iGluR extracellular domains, and more recently a full length AMPA receptor, combined with data from electrophysiological experiments and MD simulations, provides a framework that makes it possible to investigate the molecular basis for assembly, gating and modulation. These unprecedented advances in structural biology are constantly challenged by novel functional properties that emerge despite decades of functional analysis, and by a growing family of auxiliary proteins that modulate iGluR activity and assembly.
Published by Elsevier Ltd.
Figures

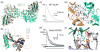
References
-
- Hollmann M. Structure of ionotropic glutamate receptors. In: Jonas P, Monyer H, editors. Ionotropic glutamate receptors in the CNS. Springer-Verlag; 1999. pp. 3–98.
- Handbook of experimental pharmacology. Vol. 141
-
- Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. - PubMed
-
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechamism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. The 1st full length crystal structure of an iGluR tetramer, which took more than 5 years to solve, confirmed many predictions made from the structures of soluble iGluR domains, but also revealed many unexpected features which have no counterpart in other neurotransmitter receptors including symmetry mismatch, large cavities, and subunit cross over between domains. - PMC - PubMed
-
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

