Serum protein profiling of systemic lupus erythematosus and systemic sclerosis using recombinant antibody microarrays
- PMID: 21350050
- PMCID: PMC3098590
- DOI: 10.1074/mcp.M110.005033
Serum protein profiling of systemic lupus erythematosus and systemic sclerosis using recombinant antibody microarrays
Abstract
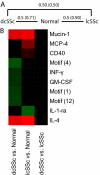
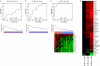
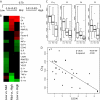
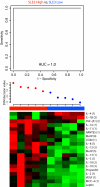
Systemic lupus erythematosus (SLE) and systemic sclerosis (SSc) are two severe autoimmune connective tissue diseases. The fundamental knowledge about their etiology is limited and the conditions display complex pathogenesis, multifaceted presentations, and unpredictable courses. Despite significant efforts, the lack of fully validated biomarkers enabling diagnosis, classification, and monitoring of disease activity represents significant unmet clinical needs. In this discovery study, we have for the first time used recombinant antibody microarrays for miniaturized, multiplexed serum protein profiling of SLE and SSc, targeting mainly immunoregulatory proteins. The data showed that several candidate SLE-associated multiplexed serum biomarker signatures were delineated, reflecting disease (diagnosis), disease severity (phenotypic subsets), and disease activity. Selected differentially expressed markers were validated using orthogonal assays and a second, independent patient cohort. Further, biomarker signatures differentiating SLE versus SSc were demonstrated, and the observed differences increased with severity of SLE. In contrast, the data showed that the serum profiles of SSc versus healthy controls were more similar. Hence, we have shown that affinity proteomics could be used to de-convolute crude, nonfractionated serum proteomes, extracting molecular portraits of SLE and SSc, further enhancing our fundamental understanding of these complex autoimmune conditions.
Figures



References
-
- D'Cruz D. P., Khamashta M. A., Hughes G. R. (2007) Systemic lupus erythematosus. Lancet 369, 587–596 - PubMed
-
- Rahman A., Isenberg D. A. (2008) Systemic lupus erythematosus. New Engl. J. Med. 358, 929–939 - PubMed
-
- Varga J. (2008) Systemic sclerosis: an update. Bull. NYU Hosp. Joint Diseases 66, 198–202 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

