Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli
- PMID: 21350490
- PMCID: PMC3078058
- DOI: 10.1038/nature09743
Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli
Abstract
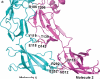
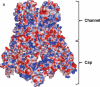
Gram-negative bacteria, such as Escherichia coli, expel toxic chemicals through tripartite efflux pumps that span both the inner and outer membrane. The three parts are an inner membrane, substrate-binding transporter; a membrane fusion protein; and an outer-membrane-anchored channel. The fusion protein connects the transporter to the channel within the periplasmic space. A crystallographic model of this tripartite efflux complex has been unavailable because co-crystallization of the various components of the system has proven to be extremely difficult. We previously described the crystal structures of both the inner membrane transporter CusA and the membrane fusion protein CusB of the CusCBA efflux system of E. coli. Here we report the co-crystal structure of the CusBA efflux complex, showing that the transporter (or pump) CusA, which is present as a trimer, interacts with six CusB protomers and that the periplasmic domain of CusA is involved in these interactions. The six CusB molecules seem to form a continuous channel. The affinity of the CusA and CusB interaction was found to be in the micromolar range. Finally, we have predicted a three-dimensional structure for the trimeric CusC outer membrane channel and developed a model of the tripartite efflux assemblage. This CusC(3)-CusB(6)-CusA(3) model shows a 750-kilodalton efflux complex that spans the entire bacterial cell envelope and exports Cu I and Ag I ions.
Figures





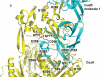
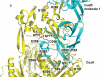


References
-
- Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiol. 2001;147:965–972. - PubMed
-
- Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development protein. J. Mol. Microbiol. Biotechnol. 1999;1:107–125. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

