CD4+ T cells from HIV-1-infected patients recognize wild-type and mutant human immunodeficiency virus-1 protease epitopes
- PMID: 21352200
- PMCID: PMC3074221
- DOI: 10.1111/j.1365-2249.2011.04319.x
CD4+ T cells from HIV-1-infected patients recognize wild-type and mutant human immunodeficiency virus-1 protease epitopes
Abstract
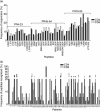
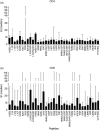
Human immunodeficiency virus (HIV)-1 protease is a known target of CD8+ T cell responses, but it is the only HIV-1 protein in which no fully characterized HIV-1 protease CD4 epitopes have been identified to date. We investigated the recognition of HIV-1 protease by CD4+ T cells from 75 HIV-1-infected, protease inhibitor (PI)-treated patients, using the 5,6-carboxyfluorescein diacetate succinimidyl ester-based proliferation assay. In order to identify putative promiscuous CD4+ T cell epitopes, we used the TEPITOPE algorithm to scan the sequence of the HXB2 HIV-1 protease. Protease regions 4-23, 45-64 and 73-95 were identified; 32 sequence variants of the mentioned regions, encoding frequent PI-induced mutations and polymorphisms, were also tested. On average, each peptide bound to five of 15 tested common human leucocyte antigen D-related (HLA-DR) molecules. More than 80% of the patients displayed CD4+ as well as CD8+ T cell recognition of at least one of the protease peptides. All 35 peptides were recognized. The response was not associated with particular HLA-DR or -DQ alleles. Our results thus indicate that protease is a frequent target of CD4+ along with CD8+ proliferative T cell responses by the majority of HIV-1-infected patients under PI therapy. The frequent finding of matching CD4(+) and CD8+ T cell responses to the same peptides may indicate that CD4+ T cells provide cognate T cell help for the maintenance of long-living protease-specific functional CD8+ T cells.
© 2011 The Authors. Clinical and Experimental Immunology © 2011 British Society for Immunology.
Figures

References
-
- Mason RD, Bowmer MI, Howley CM, et al. Antiretroviral drug resistance mutations sustain or enhance CTL recognition of common HIV-1 pol epitopes. J Immunol. 2004;172:7212–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

