Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy
- PMID: 21352579
- PMCID: PMC3058099
- DOI: 10.1186/1476-4598-10-23
Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy
Abstract
Background: Aneuploidy and chromosomal instability (CIN) are common abnormalities in human cancer. Alterations of the mitotic spindle checkpoint are likely to contribute to these phenotypes, but little is known about somatic alterations of mitotic spindle checkpoint genes in breast cancer.
Methods: To obtain further insight into the molecular mechanisms underlying aneuploidy in breast cancer, we used real-time quantitative RT-PCR to quantify the mRNA expression of 76 selected mitotic spindle checkpoint genes in a large panel of breast tumor samples.
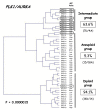
Results: The expression of 49 (64.5%) of the 76 genes was significantly dysregulated in breast tumors compared to normal breast tissues: 40 genes were upregulated and 9 were downregulated. Most of these changes in gene expression during malignant transformation were observed in epithelial cells.Alterations of nine of these genes, and particularly NDC80, were also detected in benign breast tumors, indicating that they may be involved in pre-neoplastic processes.We also identified a two-gene expression signature (PLK1 + AURKA) which discriminated between DNA aneuploid and DNA diploid breast tumor samples. Interestingly, some DNA tetraploid tumor samples failed to cluster with DNA aneuploid breast tumors.
Conclusion: This study confirms the importance of previously characterized genes and identifies novel candidate genes that could be activated for aneuploidy to occur. Further functional analyses are required to clearly confirm the role of these new identified genes in the molecular mechanisms involved in breast cancer aneuploidy. The novel genes identified here, and/or the two-gene expression signature, might serve as diagnostic or prognostic markers and form the basis for novel therapeutic strategies.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

