The Q cycle of cytochrome bc complexes: a structure perspective
- PMID: 21352799
- PMCID: PMC3101715
- DOI: 10.1016/j.bbabio.2011.02.006
The Q cycle of cytochrome bc complexes: a structure perspective
Abstract
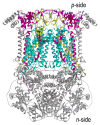
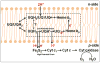
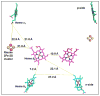
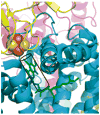
Aspects of the crystal structures of the hetero-oligomeric cytochrome bc(1) and b(6)f ("bc") complexes relevant to their electron/proton transfer function and the associated redox reactions of the lipophilic quinones are discussed. Differences between the b(6)f and bc(1) complexes are emphasized. The cytochrome bc(1) and b(6)f dimeric complexes diverge in structure from a core of subunits that coordinate redox groups consisting of two bis-histidine coordinated hemes, a heme b(n) and b(p) on the electrochemically negative (n) and positive (p) sides of the complex, the high potential [2Fe-2S] cluster and c-type heme at the p-side aqueous interface and aqueous phase, respectively, and quinone/quinol binding sites on the n- and p-sides of the complex. The bc(1) and b(6)f complexes diverge in subunit composition and structure away from this core. b(6)f Also contains additional prosthetic groups including a c-type heme c(n) on the n-side, and a chlorophyll a and β-carotene. Common structure aspects; functions of the symmetric dimer. (I) Quinone exchange with the bilayer. An inter-monomer protein-free cavity of approximately 30Å along the membrane normal×25Å (central inter-monomer distance)×15Å (depth in the center), is common to both bc(1) and b(6)f complexes, providing a niche in which the lipophilic quinone/quinol (Q/QH(2)) can be exchanged with the membrane bilayer. (II) Electron transfer. The dimeric structure and the proximity of the two hemes b(p) on the electrochemically positive side of the complex in the two monomer units allow the possibility of two alternate routes of electron transfer across the complex from heme b(p) to b(n): intra-monomer and inter-monomer involving electron cross-over between the two hemes b(p). A structure-based summary of inter-heme distances in seven bc complexes, representing mitochondrial, chromatophore, cyanobacterial, and algal sources, indicates that, based on the distance parameter, the intra-monomer pathway would be favored kinetically. (III) Separation of quinone binding sites. A consequence of the dimer structure and the position of the Q/QH(2) binding sites is that the p-side QH(2) oxidation and n-side Q reduction sites are each well separated. Therefore, in the event of an overlap in residence time by QH(2) or Q molecules at the two oxidation or reduction sites, their spatial separation would result in minimal steric interference between extended Q or QH(2) isoprenoid chains. (IV) Trans-membrane QH(2)/Q transfer. (i) n/p-side QH(2)/Q transfer may be hindered by lipid acyl chains; (ii) the shorter less hindered inter-monomer pathway across the complex would not pass through the center of the cavity, as inferred from the n-side antimycin site on one monomer and the p-side stigmatellin site on the other residing on the same surface of the complex. (V) Narrow p-side portal for QH(2)/Q passage. The [2Fe-2S] cluster that serves as oxidant, and whose histidine ligand serves as a H(+) acceptor in the oxidation of QH(2), is connected to the inter-monomer cavity by a narrow extended portal, which is also occupied in the b(6)f complex by the 20 carbon phytyl chain of the bound chlorophyll.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures















References
-
- Berry EA, Huang L-S, Earnest TN, Jap BK. X-ray diffraction by crystals of beef heart ubiquinol:cytochrome c oxidoreductase. J Mol Biol. 1992;224:1161–1166. - PubMed
-
- Xia D, Yu CA, Deisenhofer J, Xia J-Z, Yu L. Three dimensional structure of beef-heart mitochondrial cytochrome bc1 complex. Biophys J. 1996;70:253a. - PubMed
-
- Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science (New York, NY. 1998;281:64–71. - PubMed
-
- Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

