Functional identification of optimized RNAi triggers using a massively parallel sensor assay
- PMID: 21353615
- PMCID: PMC3130540
- DOI: 10.1016/j.molcel.2011.02.008
Functional identification of optimized RNAi triggers using a massively parallel sensor assay
Abstract
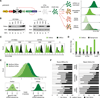
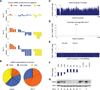
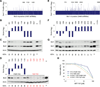
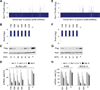
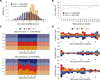
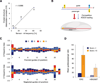
Short hairpin RNAs (shRNAs) provide powerful experimental tools by enabling stable and regulated gene silencing through programming of endogenous microRNA pathways. Since requirements for efficient shRNA biogenesis and target suppression are largely unknown, many predicted shRNAs fail to efficiently suppress their target. To overcome this barrier, we developed a "Sensor assay" that enables the biological identification of effective shRNAs at large scale. By constructing and evaluating 20,000 RNAi reporters covering every possible target site in nine mammalian transcripts, we show that our assay reliably identifies potent shRNAs that are surprisingly rare and predominantly missed by existing algorithms. Our unbiased analyses reveal that potent shRNAs share various predicted and previously unknown features associated with specific microRNA processing steps, and suggest a model for competitive strand selection. Together, our study establishes a powerful tool for large-scale identification of highly potent shRNAs and provides insights into sequence requirements of effective RNAi.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

References
-
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. - PubMed
-
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. - PubMed
-
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

