Chlorite dismutases, DyPs, and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily
- PMID: 21354424
- PMCID: PMC3075325
- DOI: 10.1016/j.jmb.2011.02.047
Chlorite dismutases, DyPs, and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily
Abstract
Heme proteins are extremely diverse, widespread, and versatile biocatalysts, sensors, and molecular transporters. The chlorite dismutase family of hemoproteins received its name due to the ability of the first-isolated members to detoxify anthropogenic ClO(2)(-), a function believed to have evolved only in the last few decades. Family members have since been found in 15 bacterial and archaeal genera, suggesting ancient roots. A structure- and sequence-based examination of the family is presented, in which key sequence and structural motifs are identified, and possible functions for family proteins are proposed. Newly identified structural homologies moreover demonstrate clear connections to two other large, ancient, and functionally mysterious protein families. We propose calling them collectively the CDE superfamily of heme proteins.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures




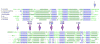
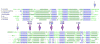



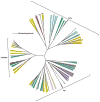
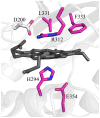


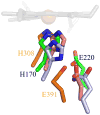




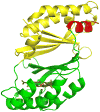


References
-
- Clark JJ. Toxicology of perchlorate. In: Urbansky ET, editor. Perchlorate in the Environment. Kluwer Academic/Plenum; New York: 2000.
-
- Hauntman DP, et al. Determination of perchlorate in drinking water using ion chromotography, revision 1 Method 314, Doc No EPA/815/B-99/003. US Environmental Protection Agency; 1999.
-
- Urbansky ET. Perchlorate as an environmental contaminant. Environ Sci Pollut Res Int. 2002;9:187–192. - PubMed
-
- Christen K. EPA perchlorate decision takes many by surprise. Environ Sci Technol. 2003;37:347A–348A. - PubMed
-
- Xu JL, Song YU, Min BK, Steinberg L, Logan BE. Microbial degradation of perchlorate: Principles and applications. Environ Eng Sci. 2003;20:405–422.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

