Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals
- PMID: 21356067
- PMCID: PMC3109589
- DOI: 10.1186/bcr2832
Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals
Abstract
Introduction: Two major high-penetrance breast cancer genes, BRCA1 and BRCA2, are responsible for approximately 20% of hereditary breast cancer (HBC) cases in Finland. Additionally, rare mutations in several other genes that interact with BRCA1 and BRCA2 increase the risk of HBC. Still, a majority of HBC cases remain unexplained which is challenging for genetic counseling. We aimed to analyze additional mutations in HBC-associated genes and to define the sensitivity of our current BRCA1/2 mutation analysis protocol used in genetic counseling.
Methods: Eighty-two well-characterized, high-risk hereditary breast and/or ovarian cancer (HBOC) BRCA1/2-founder mutation-negative Finnish individuals, were screened for germline alterations in seven breast cancer susceptibility genes, BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1. BRCA1/2 were analyzed by multiplex ligation-dependent probe amplification (MLPA) and direct sequencing. CHEK2 was analyzed by the high resolution melt (HRM) method and PALB2, RAD50, BRIP1 and CDH1 were analyzed by direct sequencing. Carrier frequencies between 82 (HBOC) BRCA1/2-founder mutation-negative Finnish individuals and 384 healthy Finnish population controls were compared by using Fisher's exact test. In silico prediction for novel missense variants effects was carried out by using Pathogenic-Or-Not -Pipeline (PON-P).
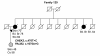
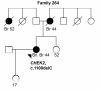
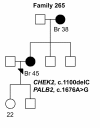
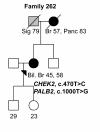
Results: Three previously reported breast cancer-associated variants, BRCA1 c.5095C > T, CHEK2 c.470T > C, and CHEK2 c.1100delC, were observed in eleven (13.4%) individuals. Ten of these individuals (12.2%) had CHEK2 variants, c.470T > C and/or c.1100delC. Fourteen novel sequence alterations and nine individuals with more than one non-synonymous variant were identified. One of the novel variants, BRCA2 c.72A > T (Leu24Phe) was predicted to be likely pathogenic in silico. No large genomic rearrangements were detected in BRCA1/2 by multiplex ligation-dependent probe amplification (MLPA).
Conclusions: In this study, mutations in previously known breast cancer susceptibility genes can explain 13.4% of the analyzed high-risk BRCA1/2-negative HBOC individuals. CHEK2 mutations, c.470T > C and c.1100delC, make a considerable contribution (12.2%) to these high-risk individuals but further segregation analysis is needed to evaluate the clinical significance of these mutations before applying them in clinical use. Additionally, we identified novel variants that warrant additional studies. Our current genetic testing protocol for 28 Finnish BRCA1/2-founder mutations and protein truncation test (PTT) of the largest exons is sensitive enough for clinical use as a primary screening tool.
Figures


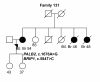

References
-
- Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, Belanger C, Bell R, Berry S, Bogden R, Chen Q, Davis T, Dumont M, Frye C, Hattier T, Jammulapati S, Janecki T, Jiang P, Kehrer R, Leblanc JF, Mitchell JT, McArthur-Morrison J, Nguyen K, Peng Y, Samson C, Schroeder M, Snyder SC. et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. - DOI - PubMed
-
- Vehmanen P, Friedman LS, Eerola H, McClure M, Ward B, Sarantaus L, Kainu T, Syrjakoski K, Pyrhonen S, Kallioniemi OP, Muhonen T, Luce M, Frank TS, Nevanlinna H. Low proportion of BRCA1 and BRCA2 mutations in Finnish breast cancer families: evidence for additional susceptibility genes. Hum Mol Genet. 1997;6:2309–2315. doi: 10.1093/hmg/6.13.2309. - DOI - PubMed
-
- Syrjakoski K, Vahteristo P, Eerola H, Tamminen A, Kivinummi K, Sarantaus L, Holli K, Blomqvist C, Kallioniemi OP, Kainu T, Nevanlinna H. Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J Natl Cancer Inst. 2000;92:1529–1531. doi: 10.1093/jnci/92.18.1529. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

