Automatically identifying and annotating mouse embryo gene expression patterns
- PMID: 21357576
- PMCID: PMC3072560
- DOI: 10.1093/bioinformatics/btr105
Automatically identifying and annotating mouse embryo gene expression patterns
Abstract
Motivation: Deciphering the regulatory and developmental mechanisms for multicellular organisms requires detailed knowledge of gene interactions and gene expressions. The availability of large datasets with both spatial and ontological annotation of the spatio-temporal patterns of gene expression in mouse embryo provides a powerful resource to discover the biological function of embryo organization. Ontological annotation of gene expressions consists of labelling images with terms from the anatomy ontology for mouse development. If the spatial genes of an anatomical component are expressed in an image, the image is then tagged with a term of that anatomical component. The current annotation is done manually by domain experts, which is both time consuming and costly. In addition, the level of detail is variable, and inevitably errors arise from the tedious nature of the task. In this article, we present a new method to automatically identify and annotate gene expression patterns in the mouse embryo with anatomical terms.
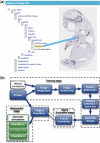
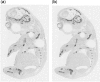
Results: The method takes images from in situ hybridization studies and the ontology for the developing mouse embryo, it then combines machine learning and image processing techniques to produce classifiers that automatically identify and annotate gene expression patterns in these images. We evaluate our method on image data from the EURExpress study, where we use it to automatically classify nine anatomical terms: humerus, handplate, fibula, tibia, femur, ribs, petrous part, scapula and head mesenchyme. The accuracy of our method lies between 70% and 80% with few exceptions. We show that other known methods have lower classification performance than ours. We have investigated the images misclassified by our method and found several cases where the original annotation was not correct. This shows our method is robust against this kind of noise.
Availability: The annotation result and the experimental dataset in the article can be freely accessed at http://www2.docm.mmu.ac.uk/STAFF/L.Han/geneannotation/.
Contact: l.han@mmu.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures



References
-
- Baldock R., et al. Emap and emage: a framework for understanding spatially organised data. Neuroinformatics. 2003;1:309–325. - PubMed
-
- Baxes G.A. Digital Image Processing: Principles and Applications. New York, NY, USA: Wiley; 1994.
-
- Brown P.O., Botstein D. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 1999;21:33–37. - PubMed

