Terminal alkene formation by the thioesterase of curacin A biosynthesis: structure of a decarboxylating thioesterase
- PMID: 21357626
- PMCID: PMC3077644
- DOI: 10.1074/jbc.M110.214635
Terminal alkene formation by the thioesterase of curacin A biosynthesis: structure of a decarboxylating thioesterase
Abstract
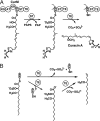
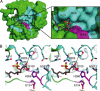
Curacin A is a polyketide synthase (PKS)-non-ribosomal peptide synthetase-derived natural product with potent anticancer properties generated by the marine cyanobacterium Lyngbya majuscula. Type I modular PKS assembly lines typically employ a thioesterase (TE) domain to off-load carboxylic acid or macrolactone products from an adjacent acyl carrier protein (ACP) domain. In a striking departure from this scheme the curacin A PKS employs tandem sulfotransferase and TE domains to form a terminal alkene moiety. Sulfotransferase sulfonation of β-hydroxy-acyl-ACP is followed by TE hydrolysis, decarboxylation, and sulfate elimination (Gu, L., Wang, B., Kulkarni, A., Gehret, J. J., Lloyd, K. R., Gerwick, L., Gerwick, W. H., Wipf, P., Håkansson, K., Smith, J. L., and Sherman, D. H. (2009) J. Am. Chem. Soc. 131, 16033-16035). With low sequence identity to other PKS TEs (<15%), the curacin TE represents a new thioesterase subfamily. The 1.7-Å curacin TE crystal structure reveals how the familiar α/β-hydrolase architecture is adapted to specificity for β-sulfated substrates. A Ser-His-Glu catalytic triad is centered in an open active site cleft between the core domain and a lid subdomain. Unlike TEs from other PKSs, the lid is fixed in an open conformation on one side by dimer contacts of a protruding helix and on the other side by an arginine anchor from the lid into the core. Adjacent to the catalytic triad, another arginine residue is positioned to recognize the substrate β-sulfate group. The essential features of the curacin TE are conserved in sequences of five other putative bacterial ACP-ST-TE tridomains. Formation of a sulfate leaving group as a biosynthetic strategy to facilitate acyl chain decarboxylation is of potential value as a route to hydrocarbon biofuels.
Figures




References
-
- Newman D. J., Cragg G. M. (2007) J. Nat. Prod. 70, 461–477 - PubMed
-
- Fortman J. L., Chhabra S., Mukhopadhyay A., Chou H., Lee T. S., Steen E., Keasling J. D. (2008) Trends Biotechnol. 26, 375–381 - PubMed
-
- Verdier-Pinard P., Lai J. Y., Yoo H. D., Yu J., Marquez B., Nagle D. G., Nambu M., White J. D., Falck J. R., Gerwick W. H., Day B. W., Hamel E. (1998) Mol. Pharmacol. 53, 62–76 - PubMed
-
- Chang Z., Sitachitta N., Rossi J. V., Roberts M. A., Flatt P. M., Jia J., Sherman D. H., Gerwick W. H. (2004) J. Nat. Prod. 67, 1356–1367 - PubMed
-
- Gu L., Geders T. W., Wang B., Gerwick W. H., Håkansson K., Smith J. L., Sherman D. H. (2007) Science 318, 970–974 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

