Autoregulation of convergent RNAi genes in fission yeast
- PMID: 21357674
- PMCID: PMC3059830
- DOI: 10.1101/gad.618611
Autoregulation of convergent RNAi genes in fission yeast
Abstract
RNAi plays a central role in the regulation of eukaryotic genes. In Schizosaccharomyces pombe fission yeast, RNAi involves the formation of siRNA from dsRNA that acts to establish and maintain heterochromatin over centromeres, telomeres, and mating loci. We showed previously that transient heterochromatin also forms over S. pombe convergent genes (CGs). Remarkably, most RNAi genes are themselves convergent. We demonstrate here that transient heterochromatin formed by the RNAi pathway over RNAi CGs leads to their autoregulation in G1-S. Furthermore, the switching of RNAi gene orientation from convergent to tandem causes loss of their G1-S down-regulation. Surprisingly, yeast mutants with tandemized dcr1, ago1, or clr4 genes display aberrant centromeric heterochromatin, which results in abnormal cell morphology. Our results emphasize the significance of gene orientation for correct RNAi gene expression, and suggest a role for cell cycle-dependent formation of RNAi CG heterochromatin in cellular integrity.
Figures




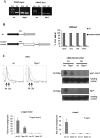
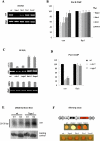

Comment in
-
RNAi genes pave their own way.Genes Dev. 2011 Mar 15;25(6):529-33. doi: 10.1101/gad.2038611. Genes Dev. 2011. PMID: 21406552 Free PMC article.
References
-
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G 1994. Position effect variegation at fission yeast centromeres. Cell 76: 157–169 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
