Up-regulation of homeodomain genes, DLX1 and DLX2, by FLT3 signaling
- PMID: 21357706
- PMCID: PMC3105643
- DOI: 10.3324/haematol.2010.031179
Up-regulation of homeodomain genes, DLX1 and DLX2, by FLT3 signaling
Abstract
Background: Activating mutations in fms-like tyrosine kinase-3 (FLT3) are frequent in acute myeloid leukemia and represent both a poor prognostic feature and a therapeutic target. We have identified a previously unrecognized downstream effect of FLT3 activation, namely up-regulation of the homeodomain genes, DLX1 and DLX2.
Design and methods: MV4;11 cells with FLT3-internal tandem duplication mutation, RS4;11 cells with wild-type FLT3 and blasts from patients with acute myeloid leukemia were used to pursue the relation between FLT3, DLX1/2 and transforming growth factor-β (TGFβ). Real-time quantitative reverse transcriptase polymerase chain reaction, western blot and reverse-phase protein array were performed to detect changes in gene and protein expression. RNA interference and MTS assays were used to study the interaction of PKC412, FLT3 inhibitor and TGFβ1.
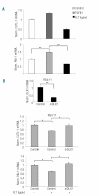
Results: A direct relationship between FLT3 activity and DLX1/2 expression was revealed by both inhibition and up-regulation of FLT3 signaling in MV4;11 and RS4;11 cell lines, respectively, in isolated blast cells from patients with acute myeloid leukemia, and in reverse-phase protein array assays of samples from patients with acute myeloid leukemia. Mechanistically, the link between FLT3 and DLX1 expression appears to involve MAPK signaling through the ERK and JNK pathways. To determine whether elevated DLX1 had a functional consequence, we explored the reported inhibition by DLX1 on TGFβ/Smad signaling. Indeed, TGFβ responses were blunted by FLT3 activation in a DLX1-dependent manner and FLT3 inhibition resulted in a time-dependent increase in nuclear phospho-Smad2.
Conclusions: These findings suggest that alterations in DLX1/2 contribute to the biological consequences of FLT3 activation.
Figures
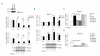




References
-
- Apiou F, Flagiello D, Cillo C, Malfoy B, Poupon MF, Dutrillaux B. Fine mapping of human HOX gene clusters. Cytogenet Cell Genet. 1996;73(1–2):114–5. - PubMed
-
- Fujino T, Suzuki A, Ito Y, Ohyashiki K, Hatano Y, Miura I, et al. Single-translocation and double-chimeric transcripts: detection of NUP98-HOXA9 in myeloid leukemias with HOXA11 or HOXA13 breaks of the chromosomal translocation t(7;11)(p15;p15) Blood. 2002;99(4):1428–33. - PubMed
-
- Lahortiga I, Belloni E, Vazquez I, Agirre X, Larrayoz MJ, Vizmanos JL, et al. NUP98 is fused to HOXA9 in a variant complex t(7;11;13;17) in a patient with AML-M2. Cancer Genet Cytogenet. 2005;157 (2):151–6. - PubMed
-
- Nakamura T. NUP98 fusion in human leukemia: dysregulation of the nuclear pore and homeodomain proteins. Int J Hematol. 2005;82(1):21–7. - PubMed
-
- Nishiyama M, Arai Y, Tsunematsu Y, Kobayashi H, Asami K, Yabe M, et al. 11p15 translocations involving the NUP98 gene in childhood therapy-related acute myeloid leukemia/myelodysplastic syndrome. Genes Chromosomes Cancer. 1999;26(3):215–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

