Elongator protein 3b negatively regulates ribosomal DNA transcription in african trypanosomes
- PMID: 21357738
- PMCID: PMC3133226
- DOI: 10.1128/MCB.01026-10
Elongator protein 3b negatively regulates ribosomal DNA transcription in african trypanosomes
Abstract
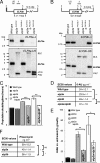
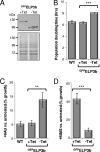
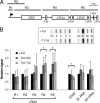
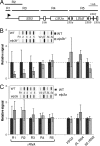
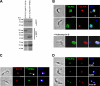
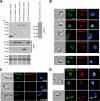
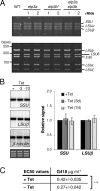
Eukaryotic cells limit ribosomal DNA (rDNA) transcription by RNA polymerase I (RNAP-I) to maintain genome integrity. African trypanosomes present an excellent model for studies on RNAP-I regulation because they possess a bifunctional RNAP-I and because RNAP-II transcription appears unregulated. Since Elp3, the catalytic component of Elongator, controls RNAP-II transcription in yeast and human cells, we predicted a role for a trypanosome Elp3-related protein, ELP3a or ELP3b, in RNAP-I regulation. elp3b null and conditional strains specifically exhibited resistance to a transcription elongation inhibitor, suggesting that ELP3b negatively impacts elongation. Nascent RNA analysis and expression of integrated reporter cassettes supported this interpretation and revealed negative control of rDNA transcription. ELP3b specifically localized to the nucleolus, and ELP3b loss rendered cells hypersensitive to DNA damage and to translation inhibition, suggesting that anti-Elongator function was important to maintain genome integrity rather than to modulate ribosome production. Finally, ELP3b displayed discrimination between RNAP-I compartments in the same cell. Our results establish ELP3b as a major negative regulator of rDNA transcription and extend the roles of the Elp3-related proteins to RNAP-I transcription units. ELP3b is also the first trypanosome protein shown to distinguish between rDNA and variant surface glycoprotein transcription within different RNAP-I compartments.
Figures


References
-
- Alibu V. P., Storm L., Haile S., Clayton C., Horn D. 2005. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:75–82 - PubMed
-
- Alsford S., Kawahara T., Isamah C., Horn D. 2007. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol. Microbiol. 63:724–736 - PubMed
-
- Ausubel F. M., et al. 1998. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
