T-body formation precedes virus-like particle maturation in S. cerevisiae
- PMID: 21358276
- PMCID: PMC3127098
- DOI: 10.4161/rna.8.2.14822
T-body formation precedes virus-like particle maturation in S. cerevisiae
Abstract
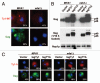
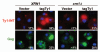
T-bodies are localized S. cerevisiae RNPs containing Ty1 retroviral components and speculated to play a role in the assembly of virus-like particles (VLPs). Mapping requirements for T-body formation, we demonstrate that ectopic expression of immature TyA1/Gag (Gag-p49), a structural component of the Ty1 capsid, is sufficient for T-body formation both under normal conditions as well as in a strain background devoid of endogenous Gag. Moreover, T-bodies are readily formed when Ty1 transposition is blocked. Thus, T-bodies represent an early stage in the Ty1 life cycle, preceding VLP maturation.
Figures


Similar articles
-
The Ty1 Retrotransposon Restriction Factor p22 Targets Gag.PLoS Genet. 2015 Oct 9;11(10):e1005571. doi: 10.1371/journal.pgen.1005571. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26451601 Free PMC article.
-
A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein.J Virol. 1996 Aug;70(8):5548-56. doi: 10.1128/JVI.70.8.5548-5556.1996. J Virol. 1996. PMID: 8764068 Free PMC article.
-
Possible regulatory function of the Saccharomyces cerevisiae Ty1 retrotransposon core protein.Yeast. 2000 Jul;16(10):921-32. doi: 10.1002/1097-0061(200007)16:10<921::AID-YEA588>3.0.CO;2-#. Yeast. 2000. PMID: 10870103
-
The yeast Ty virus-like particles.Yeast. 2000 Jun 30;16(9):785-95. doi: 10.1002/1097-0061(20000630)16:9<785::AID-YEA550>3.0.CO;2-L. Yeast. 2000. PMID: 10861903 Review.
-
A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces.Curr Genet. 2016 May;62(2):321-9. doi: 10.1007/s00294-015-0550-6. Epub 2015 Dec 9. Curr Genet. 2016. PMID: 26650614 Free PMC article. Review.
Cited by
-
Mapping the structural landscape of the yeast Ty3 retrotransposon RNA genome.Nucleic Acids Res. 2024 Sep 9;52(16):9821-9837. doi: 10.1093/nar/gkae494. Nucleic Acids Res. 2024. PMID: 38864374 Free PMC article.
-
Preferential retrotransposition in aging yeast mother cells is correlated with increased genome instability.DNA Repair (Amst). 2015 Oct;34:18-27. doi: 10.1016/j.dnarep.2015.07.004. Epub 2015 Aug 7. DNA Repair (Amst). 2015. PMID: 26298836 Free PMC article.
-
A purine loop and the primer binding site are critical for the selective encapsidation of mouse mammary tumor virus genomic RNA by Pr77Gag.Nucleic Acids Res. 2021 May 7;49(8):4668-4688. doi: 10.1093/nar/gkab223. Nucleic Acids Res. 2021. PMID: 33836091 Free PMC article.
-
Characterizing the functions of Ty1 Gag and the Gag-derived restriction factor p22/p18.Mob Genet Elements. 2016 Mar 7;6(2):e1154637. doi: 10.1080/2159256X.2016.1154637. eCollection 2016 Mar-Apr. Mob Genet Elements. 2016. PMID: 27141325 Free PMC article.
-
The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr 1;3(2):1-35. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 25893143 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
