Enhancer function: new insights into the regulation of tissue-specific gene expression
- PMID: 21358745
- PMCID: PMC3175006
- DOI: 10.1038/nrg2957
Enhancer function: new insights into the regulation of tissue-specific gene expression
Abstract
Enhancer function underlies regulatory processes by which cells establish patterns of gene expression. Recent results suggest that many enhancers are specified by particular chromatin marks in pluripotent cells, which may be modified later in development to alter patterns of gene expression and cell differentiation choices. These marks may contribute to the repertoire of epigenetic mechanisms responsible for cellular memory and determine the timing of transcription factor accessibility to the enhancer. Mechanistically, cohesin and non-coding RNAs are emerging as crucial players responsible for facilitating enhancer-promoter interactions at some genes. Surprisingly, these interactions may be required not only to facilitate initiation of transcription but also to activate the release of RNA polymerase II (RNAPII) from promoter-proximal pausing.
Figures

| CBP: | CREB (Cyclic-AMP Response Element Binding)-binding protein. |
| CTCF: | CCCTC-binding factor. |
| DPE: | Distal promoter elements. |
| H3K4me1/2: | histone H3 mono/di-methylation at lysine 4. |
| H3K4me3: | histone H3 trimethylation at lysine 4. |
| H3K27me3: | histone H3 trimethylation at lysine 27. |
| H3.3/H2A.Z: | histone variants H3.3 and H2A.Z. |
| LCR: | Locus control region. |
| RNAPII: | RNA polymerase II. |
| TATA: | 5′-TATAAAA-3′ core DNA sequences. |
| TSS: | Transcription start site. |

| AR: | Androgen receptor. |
| CD4: | Cluster of differentiation 4. |
| DHT: | Dihydrotestosterone. |
| E47: | Immunoglobulin E2-Box Binding Protein isoform 47. |
| FoxA1: | Foxhead box A1. |
| H3/H2: | histone H3 and H2. |
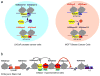
| AR: | Androgen receptor. |
| ER: | Estrogen receptor. |
| FoxA1: | Foxhead box A1. |
| Foxd3: | Foxhead box D3. |
| H3K9me2: | histone H3 dimethylation at lysine 9. |
| Sox2: | SRY (sex determining region Y)-box 2. |

| APO: | Apolipoprotein. |
| CTCF: | CCCTC-binding factor. |
| IL26: | Interleukins 26. |
| IFNγ: | Interferon-gamma. |
| Oct4: | octamer-binding transcription factor 4. |
| RNAPII: | RNA polymerase II. |
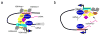
| Arc: | Activity-regulated cytoskeleton-associated protein |
| CREB: | Cyclic-AMP Response Element Binding Protein. |
| CBP: | CREB-binding protein. |
| eRNA: | enhancer RNA. |
| mRNA: | messenger RNA. |
| ncRNA: | non-coding RNA. |
| Snai1: | Snail homolog 1a. |

| Brd4: | Bromodomain containing 4 |
| ELP-3: | Elongator complex protein 3. |
| FOSL1: | FOS-like antigen 1. |
| H3K9acS10P: | histone H3 acetylation at lysine 9 and phosphorylation at serine 10. |
| H4K16ac: | histone H4 acetylation at lysine 16. |
| MOF: | Male absent on the first, histone H4 lysine 16-specific acetyltransferase. |
| MSK: | Mitogen and Stress-Activated Protein Kinase. |
| PIM1: | Proviral integration site 1, a proto-oncogene serine/threonine-protein kinase. |
| P-TEFb: | positive transcription elongation factor b (P-TEFb). |
| RNAPII: | RNA polymerase II. |
References
-
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. - PubMed
-
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. - PubMed
-
- Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

