Strategies for the discovery of therapeutic aptamers
- PMID: 21359096
- PMCID: PMC3045091
- DOI: 10.1517/17460441.2011.537321
Strategies for the discovery of therapeutic aptamers
Abstract
Importance of the field: Therapeutic aptamers are synthetic, structured oligonucleotides that bind to a very broad range of targets with high affinity and specificity. They are an emerging class of targeting ligand that show great promise for treating a number of diseases. A series of aptamers currently in various stages of clinical development highlights the potential of aptamers for therapeutic applications.
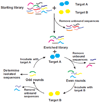
Areas covered in this review: This review covers in vitro selection of oligonucleotide ligands, called aptamers, from a combinatorial library using the Systematic Evolution of Ligands by Exponential Enrichment process as well as the other known strategies for finding aptamers against various targets.
What the reader will gain: Readers will gain an understanding of the highly useful strategies for successful aptamer discovery. They may also be able to combine two or more of the presented strategies for their aptamer discovery projects.
Take home message: Although many processes are available for discovering aptamers, it is not easy to discover an aptamer candidate that is ready to move toward pharmaceutical drug development. It is also apparent that there have been relatively few therapeutic advances and clinical trials undertaken due to the small number of companies that participate in aptamer development.
Keywords: SELEX; aptamers; in vitro selection; oligonucleotide phosphorodithioate; thioaptamer.
Figures



Similar articles
-
Aptamers as imaging agents.Expert Opin Med Diagn. 2010 Nov;4(6):511-8. doi: 10.1517/17530059.2010.516248. Epub 2010 Sep 27. Expert Opin Med Diagn. 2010. PMID: 23496229
-
[Aptamers in the Treatment of Bacterial Infections: Problems and Prospects].Vestn Ross Akad Med Nauk. 2016;71(5):350-8. doi: 10.15690/vramn591. Vestn Ross Akad Med Nauk. 2016. PMID: 29297663 Review. Russian.
-
Aptamers 101: aptamer discovery and in vitro applications in biosensors and separations.Chem Sci. 2023 May 2;14(19):4961-4978. doi: 10.1039/d3sc00439b. eCollection 2023 May 17. Chem Sci. 2023. PMID: 37206388 Free PMC article. Review.
-
DNA and RNA aptamers: from tools for basic research towards therapeutic applications.Comb Chem High Throughput Screen. 2006 Sep;9(8):619-32. doi: 10.2174/138620706778249695. Comb Chem High Throughput Screen. 2006. PMID: 17017882 Review.
-
Recent progress of SELEX methods for screening nucleic acid aptamers.Talanta. 2024 Jan 1;266(Pt 1):124998. doi: 10.1016/j.talanta.2023.124998. Epub 2023 Jul 26. Talanta. 2024. PMID: 37527564 Review.
Cited by
-
Therapeutic Potential of Aptamer-Protein Interactions.ACS Pharmacol Transl Sci. 2022 Nov 4;5(12):1211-1227. doi: 10.1021/acsptsci.2c00156. eCollection 2022 Dec 9. ACS Pharmacol Transl Sci. 2022. PMID: 36524009 Free PMC article. Review.
-
Aptamers and the next generation of diagnostic reagents.Proteomics Clin Appl. 2012 Dec;6(11-12):563-73. doi: 10.1002/prca.201200042. Proteomics Clin Appl. 2012. PMID: 23090891 Free PMC article. Review.
-
A novel DNA aptamer targeting lung cancer stem cells exerts a therapeutic effect by binding and neutralizing Annexin A2.Mol Ther Nucleic Acids. 2022 Jan 19;27:956-968. doi: 10.1016/j.omtn.2022.01.012. eCollection 2022 Mar 8. Mol Ther Nucleic Acids. 2022. PMID: 35211356 Free PMC article.
-
In vitro selection of DNA aptamers against human osteosarcoma.Invest New Drugs. 2022 Feb;40(1):172-181. doi: 10.1007/s10637-021-01161-y. Epub 2021 Aug 12. Invest New Drugs. 2022. PMID: 34383183
-
Aptamer nucleotide analog drug conjugates in the targeting therapy of cancers.Front Cell Dev Biol. 2022 Dec 5;10:1053984. doi: 10.3389/fcell.2022.1053984. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36544906 Free PMC article. Review.
References
-
- Paul SM, Mytelka DS, Dunwiddie CT, et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9:203–214. - PubMed
-
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. - PubMed
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. - PubMed
-
- Bassett SE, Fennewald SM, King DJ, et al. Combinatorial selection and edited combinatorial selection of phosphorothioate aptamers targeting human nuclear factor-kappaB RelA/p50 and RelA/RelA. Biochemistry. 2004;43:9105–9115. - PubMed
-
- King DJ, Ventura DA, Brasier AR, et al. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37:16489–16493. - PubMed
Publication types
Grants and funding
- HHSN272200800048C/AI/NIAID NIH HHS/United States
- N01-HV-28184/HV/NHLBI NIH HHS/United States
- GM 084552/GM/NIGMS NIH HHS/United States
- RC2GM092599ARRA/GM/NIGMS NIH HHS/United States
- RC2 GM092599/GM/NIGMS NIH HHS/United States
- N01 HV028184/HL/NHLBI NIH HHS/United States
- HHSN 272200800048C/PHS HHS/United States
- N01 HV028184/HV/NHLBI NIH HHS/United States
- AI 27744/AI/NIAID NIH HHS/United States
- R01 AI027744/AI/NIAID NIH HHS/United States
- R44 GM084552/GM/NIGMS NIH HHS/United States
- R01CA128797/CA/NCI NIH HHS/United States
- R43 GM084552/GM/NIGMS NIH HHS/United States
- R01 CA128797/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
