Secretion of novel SEL1L endogenous variants is promoted by ER stress/UPR via endosomes and shed vesicles in human cancer cells
- PMID: 21359144
- PMCID: PMC3040770
- DOI: 10.1371/journal.pone.0017206
Secretion of novel SEL1L endogenous variants is promoted by ER stress/UPR via endosomes and shed vesicles in human cancer cells
Abstract
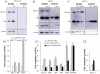
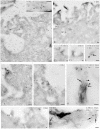
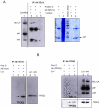
We describe here two novel endogenous variants of the human endoplasmic reticulum (ER) cargo receptor SEL1LA, designated p38 and p28. Biochemical and RNA interference studies in tumorigenic and non-tumorigenic cells indicate that p38 and p28 are N-terminal, ER-anchorless and more stable relative to the canonical transmembrane SEL1LA. P38 is expressed and constitutively secreted, with increase after ER stress, in the KMS11 myeloma line and in the breast cancer lines MCF7 and SKBr3, but not in the non-tumorigenic breast epithelial MCF10A line. P28 is detected only in the poorly differentiated SKBr3 cell line, where it is secreted after ER stress. Consistently with the presence of p38 and p28 in culture media, morphological studies of SKBr3 and KMS11 cells detect N-terminal SEL1L immunolabeling in secretory/degradative compartments and extracellularly-released membrane vesicles. Our findings suggest that the two new SEL1L variants are engaged in endosomal trafficking and secretion via vesicles, which could contribute to relieve ER stress in tumorigenic cells. P38 and p28 could therefore be relevant as diagnostic markers and/or therapeutic targets in cancer.
Conflict of interest statement
Figures



References
-
- Reed SI. The ubiquitin-proteasome pathway in cell cycle control. Results Probl Cell Differ. 2006;42:147–181. - PubMed
-
- Bader N, Jung T, Grune T. The proteasome and its role in nuclear protein maintenance. Exp Gerontol. 2007;42:864–70. - PubMed
-
- Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–19. - PubMed
-
- Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death and Differentiation. 2004;11:372–380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

