Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei
- PMID: 21359640
- PMCID: PMC3123455
- DOI: 10.1007/s10969-011-9102-6
Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei
Abstract
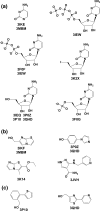
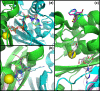
As part of the Seattle Structural Genomics Center for Infectious Disease, we seek to enhance structural genomics with ligand-bound structure data which can serve as a blueprint for structure-based drug design. We have adapted fragment-based screening methods to our structural genomics pipeline to generate multiple ligand-bound structures of high priority drug targets from pathogenic organisms. In this study, we report fragment screening methods and structure determination results for 2C-methyl-D-erythritol-2,4-cyclo-diphosphate (MECP) synthase from Burkholderia pseudomallei, the gram-negative bacterium which causes melioidosis. Screening by nuclear magnetic resonance spectroscopy as well as crystal soaking followed by X-ray diffraction led to the identification of several small molecules which bind this enzyme in a critical metabolic pathway. A series of complex structures obtained with screening hits reveal distinct binding pockets and a range of small molecules which form complexes with the target. Additional soaks with these compounds further demonstrate a subset of fragments to only bind the protein when present in specific combinations. This ensemble of fragment-bound complexes illuminates several characteristics of MECP synthase, including a previously unknown binding surface external to the catalytic active site. These ligand-bound structures now serve to guide medicinal chemists and structural biologists in rational design of novel inhibitors for this enzyme.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

