Reconstruction of brachytherapy seed positions and orientations from cone-beam CT x-ray projections via a novel iterative forward projection matching method
- PMID: 21361216
- PMCID: PMC3036175
- DOI: 10.1118/1.3528220
Reconstruction of brachytherapy seed positions and orientations from cone-beam CT x-ray projections via a novel iterative forward projection matching method
Abstract
Purpose: To generalize and experimentally validate a novel algorithm for reconstructing the 3D pose (position and orientation) of implanted brachytherapy seeds from a set of a few measured 2D cone-beam CT (CBCT) x-ray projections.
Methods: The iterative forward projection matching (IFPM) algorithm was generalized to reconstruct the 3D pose, as well as the centroid, of brachytherapy seeds from three to ten measured 2D projections. The gIFPM algorithm finds the set of seed poses that minimizes the sum-of-squared-difference of the pixel-by-pixel intensities between computed and measured autosegmented radiographic projections of the implant. Numerical simulations of clinically realistic brachytherapy seed configurations were performed to demonstrate the proof of principle. An in-house machined brachytherapy phantom, which supports precise specification of seed position and orientation at known values for simulated implant geometries, was used to experimentally validate this algorithm. The phantom was scanned on an ACUITY CBCT digital simulator over a full 660 sinogram projections. Three to ten x-ray images were selected from the full set of CBCT sinogram projections and postprocessed to create binary seed-only images.
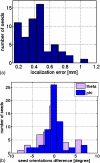
Results: In the numerical simulations, seed reconstruction position and orientation errors were approximately 0.6 mm and 5 degrees, respectively. The physical phantom measurements demonstrated an absolute positional accuracy of (0.78 +/- 0.57) mm or less. The theta and phi angle errors were found to be (5.7 +/- 4.9) degrees and (6.0 +/- 4.1) degrees, respectively, or less when using three projections; with six projections, results were slightly better. The mean registration error was better than 1 mm/6 degrees compared to the measured seed projections. Each test trial converged in 10-20 iterations with computation time of 12-18 min/iteration on a 1 GHz processor.
Conclusions: This work describes a novel, accurate, and completely automatic method for reconstructing seed orientations, as well as centroids, from a small number of radiographic projections, in support of intraoperative planning and adaptive replanning. Unlike standard back-projection methods, gIFPM avoids the need to match corresponding seed images on the projections. This algorithm also successfully reconstructs overlapping clustered and highly migrated seeds in the implant. The accuracy of better than 1 mm and 6 degrees demonstrates that gIFPM has the potential to support 2D Task Group 43 calculations in clinical practice.
Figures










Similar articles
-
Localizing intracavitary brachytherapy applicators from cone-beam CT x-ray projections via a novel iterative forward projection matching algorithm.Med Phys. 2011 Feb;38(2):1070-80. doi: 10.1118/1.3544661. Med Phys. 2011. PMID: 21452744 Free PMC article.
-
Clinical application and validation of an iterative forward projection matching algorithm for permanent brachytherapy seed localization from conebeam-CT x-ray projections.Med Phys. 2010 Sep;37(9):5092-101. doi: 10.1118/1.3480962. Med Phys. 2010. PMID: 20964229 Free PMC article.
-
Prostate brachytherapy seed localization by analysis of multiple projections: identifying and addressing the seed overlap problem.Med Phys. 2004 May;31(5):1277-87. doi: 10.1118/1.1707740. Med Phys. 2004. PMID: 15191320 Clinical Trial.
-
A Survey of the Use of Iterative Reconstruction Algorithms in Electron Microscopy.Biomed Res Int. 2017;2017:6482567. doi: 10.1155/2017/6482567. Epub 2017 Sep 17. Biomed Res Int. 2017. PMID: 29312997 Free PMC article. Review.
-
A review of brachytherapy physical phantoms developed over the last 20 years: clinical purpose and future requirements.J Contemp Brachytherapy. 2021 Feb;13(1):101-115. doi: 10.5114/jcb.2021.103593. Epub 2021 Feb 18. J Contemp Brachytherapy. 2021. PMID: 34025743 Free PMC article. Review.
Cited by
-
Automatic seed picking for brachytherapy postimplant validation with 3D CT images.Int J Comput Assist Radiol Surg. 2017 Nov;12(11):1985-1993. doi: 10.1007/s11548-017-1632-3. Epub 2017 Jun 22. Int J Comput Assist Radiol Surg. 2017. PMID: 28643024
-
Localizing intracavitary brachytherapy applicators from cone-beam CT x-ray projections via a novel iterative forward projection matching algorithm.Med Phys. 2011 Feb;38(2):1070-80. doi: 10.1118/1.3544661. Med Phys. 2011. PMID: 21452744 Free PMC article.
-
Prostate implant reconstruction from C-arm images with motion-compensated tomosynthesis.Med Phys. 2011 Oct;38(10):5290-302. doi: 10.1118/1.3633897. Med Phys. 2011. PMID: 21992346 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

