Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression
- PMID: 21362550
- PMCID: PMC3062258
- DOI: 10.1016/j.molcel.2011.02.006
Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression
Abstract
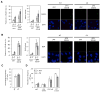
Eukaryotic cells license far more origins than are actually used for DNA replication, thereby generating a large number of dormant origins. Accumulating evidence suggests that such origins play a role in chromosome stability and tumor suppression, though the underlying mechanism is largely unknown. Here, we show that a loss of dormant origins results in an increased number of stalled replication forks, even in unchallenged S phase in primary mouse fibroblasts derived from embryos homozygous for the Mcm4(Chaos3) allele. We found that this allele reduces the stability of the MCM2-7 complex, but confers normal helicase activity in vitro. Despite the activation of multiple fork recovery pathways, replication intermediates in these cells persist into M phase, increasing the number of abnormal anaphase cells with lagging chromosomes and/or acentric fragments. These findings suggest that dormant origins constitute a major pathway for stalled fork recovery, contributing to faithful chromosome segregation and tumor suppression.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


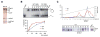


Comment in
-
A role for dormant origins in tumor suppression.Mol Cell. 2011 Mar 4;41(5):495-6. doi: 10.1016/j.molcel.2011.02.014. Mol Cell. 2011. PMID: 21362544 Free PMC article.
References
-
- Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, Chen SM, Abraham RT, Wang XF. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature. 2001;411:969–974. - PubMed
-
- Blackwell BN, Bucci TJ, Hart RW, Turturro A. Longevity, body weight, and neoplasia in ad libitum-fed and diet-restricted C57BL6 mice fed NIH-31 open formula diet. Toxicol Pathol. 1995;23:570–582. - PubMed
-
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

