A specific two-pore domain potassium channel blocker defines the structure of the TASK-1 open pore
- PMID: 21362619
- PMCID: PMC3077598
- DOI: 10.1074/jbc.M111.227884
A specific two-pore domain potassium channel blocker defines the structure of the TASK-1 open pore
Abstract
Two-pore domain potassium (K(2P)) channels play a key role in setting the membrane potential of excitable cells. Despite their role as putative targets for drugs and general anesthetics, little is known about the structure and the drug binding site of K(2P) channels. We describe A1899 as a potent and highly selective blocker of the K(2P) channel TASK-1. As A1899 acts as an open-channel blocker and binds to residues forming the wall of the central cavity, the drug was used to further our understanding of the channel pore. Using alanine mutagenesis screens, we have identified residues in both pore loops, the M2 and M4 segments, and the halothane response element to form the drug binding site of TASK-1. Our experimental data were used to validate a K(2P) open-pore homology model of TASK-1, providing structural insights for future rational design of drugs targeting K(2P) channels.
Figures
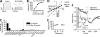

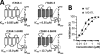


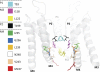


References
-
- Rajan S., Wischmeyer E., Xin Liu G., Preisig-Müller R., Daut J., Karschin A., Derst C. (2000) J. Biol. Chem. 275, 16650–16657 - PubMed
-
- Ashmole I., Goodwin P. A., Stanfield P. R. (2001) Pflügers Arch. 442, 828–833 - PubMed
-
- Decher N., Maier M., Dittrich W., Gassenhuber J., Brüggemann A., Busch A. E., Steinmeyer K. (2001) FEBS Lett. 492, 84–89 - PubMed
-
- Reyes R., Duprat F., Lesage F., Fink M., Salinas M., Farman N., Lazdunski M. (1998) J. Biol. Chem. 273, 30863–30869 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

