Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications
- PMID: 21363960
- PMCID: PMC3049283
- DOI: 10.1101/gad.2021311
Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications
Abstract
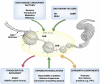
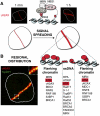
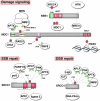
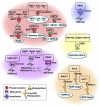
Genome integrity is constantly monitored by sophisticated cellular networks, collectively termed the DNA damage response (DDR). A common feature of DDR proteins is their mobilization in response to genotoxic stress. Here, we outline how the development of various complementary methodologies has provided valuable insights into the spatiotemporal dynamics of DDR protein assembly/disassembly at sites of DNA strand breaks in eukaryotic cells. Considerable advances have also been made in understanding the underlying molecular mechanisms for these events, with post-translational modifications of DDR factors being shown to play prominent roles in controlling the formation of foci in response to DNA-damaging agents. We review these regulatory mechanisms and discuss their biological significance to the DDR.
Figures


References
-
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451: 81–85 - PubMed
-
- Alpi AF, Patel KJ 2009. Monoubiquitylation in the Fanconi anemia DNA damage response pathway. DNA Repair (Amst) 8: 430–435 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
