The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice
- PMID: 21364282
- PMCID: PMC3069782
- DOI: 10.1172/JCI44883
The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice
Abstract
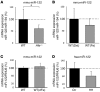
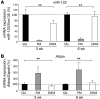
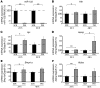
Systemic iron homeostasis is mainly controlled by the liver through synthesis of the peptide hormone hepcidin (encoded by Hamp), the key regulator of duodenal iron absorption and macrophage iron release. Here we show that the liver-specific microRNA miR-122 is important for regulating Hamp mRNA expression and tissue iron levels. Efficient and specific depletion of miR-122 by injection of a locked-nucleic-acid-modified (LNA-modified) anti-miR into WT mice caused systemic iron deficiency, characterized by reduced plasma and liver iron levels, mildly impaired hematopoiesis, and increased extramedullary erythropoiesis in the spleen. Moreover, miR-122 inhibition increased the amount of mRNA transcribed by genes that control systemic iron levels, such as hemochromatosis (Hfe), hemojuvelin (Hjv), bone morphogenetic protein receptor type 1A (Bmpr1a), and Hamp. Importantly, miR-122 directly targeted the 3′ untranslated region of 2 mRNAs that encode activators of hepcidin expression, Hfe and Hjv. These data help to explain the increased Hamp mRNA levels and subsequent iron deficiency in mice with reduced miR-122 levels and establish a direct mechanistic link between miR-122 and the regulation of systemic iron metabolism.
Figures
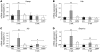

Comment in
-
The control of iron homeostasis: microRNAS join the party.Gastroenterology. 2011 Oct;141(4):1520-2. doi: 10.1053/j.gastro.2011.08.018. Epub 2011 Aug 26. Gastroenterology. 2011. PMID: 21872594 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

