An automated fluorescence videomicroscopy assay for the detection of mitotic catastrophe
- PMID: 21364633
- PMCID: PMC3032329
- DOI: 10.1038/cddis.2010.6
An automated fluorescence videomicroscopy assay for the detection of mitotic catastrophe
Abstract
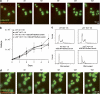
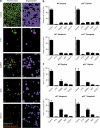
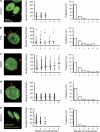
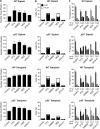
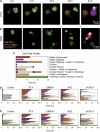
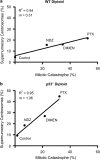
Mitotic catastrophe can be defined as a cell death mode that occurs during or shortly after a prolonged/aberrant mitosis, and can show apoptotic or necrotic features. However, conventional procedures for the detection of apoptosis or necrosis, including biochemical bulk assays and cytofluorometric techniques, cannot discriminate among pre-mitotic, mitotic and post-mitotic death, and hence are inappropriate to monitor mitotic catastrophe. To address this issue, we generated isogenic human colon carcinoma cell lines that differ in ploidy and p53 status, yet express similar amounts of fluorescent biosensors that allow for the visualization of chromatin (histone H2B coupled to green fluorescent protein (GFP)) and centrosomes (centrin coupled to the Discosoma striata red fluorescent protein (DsRed)). By combining high-resolution fluorescence videomicroscopy and automated image analysis, we established protocols and settings for the simultaneous assessment of ploidy, mitosis, centrosome number and cell death (which in our model system occurs mainly by apoptosis). Time-lapse videomicroscopy showed that this approach can be used for the high-throughput detection of mitotic catastrophe induced by three mechanistically distinct anti-mitotic agents (dimethylenastron (DIMEN), nocodazole (NDZ) and paclitaxel (PTX)), and - in this context - revealed an important role of p53 in the control of centrosome number.
Figures

References
-
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. - PubMed
-
- Galluzzi L, Morselli E, Kepp O, Kroemer G. Targeting post-mitochondrial effectors of apoptosis for neuroprotection. Biochim Biophys Acta. 2009;1787:402–413. - PubMed
-
- Galluzzi L, Blomgren K, Kroemer G. Mitochondrial membrane permeabilization in neuronal injury. Nat Rev Neurosci. 2009;10:481–494. - PubMed
-
- Galluzzi L, Larochette N, Zamzami N, Kroemer G. Mitochondria as therapeutic targets for cancer chemotherapy. Oncogene. 2006;25:4812–4830. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

