Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction
- PMID: 21364763
- PMCID: PMC3045454
- DOI: 10.1371/journal.pone.0017412
Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction
Abstract
Autophagy is a highly-conserved cellular degradation and recycling system that is essential for cell survival during nutrient starvation. The loss of viability had been used as an initial screen to identify autophagy-defective (atg) mutants of the yeast Saccharomyces cerevisiae, but the mechanism of cell death in these mutants has remained unclear. When cells grown in a rich medium were transferred to a synthetic nitrogen starvation media, secreted metabolites lowered the extracellular pH below 3.0 and autophagy-defective mutants mostly died. We found that buffering of the starvation medium dramatically restored the viability of atg mutants. In response to starvation, wild-type (WT) cells were able to upregulate components of the respiratory pathway and ROS (reactive oxygen species) scavenging enzymes, but atg mutants lacked this synthetic capacity. Consequently, autophagy-defective mutants accumulated the high level of ROS, leading to deficient respiratory function, resulting in the loss of mitochondria DNA (mtDNA). We also showed that mtDNA deficient cells are subject to cell death under low pH starvation conditions. Taken together, under starvation conditions non-selective autophagy, rather than mitophagy, plays an essential role in preventing ROS accumulation, and thus in maintaining mitochondria function. The failure of response to starvation is the major cause of cell death in atg mutants.
Conflict of interest statement
Figures


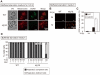
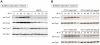
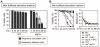
References
-
- Kornitzer D, Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol. 2000;182:1–11. - PubMed
-
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. - PubMed
-
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. - PubMed
-
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. - PubMed
-
- Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12:1535–1541. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

