Genome-wide association study identifies single nucleotide polymorphism in DYRK1A associated with replication of HIV-1 in monocyte-derived macrophages
- PMID: 21364930
- PMCID: PMC3045405
- DOI: 10.1371/journal.pone.0017190
Genome-wide association study identifies single nucleotide polymorphism in DYRK1A associated with replication of HIV-1 in monocyte-derived macrophages
Abstract
Background: HIV-1 infected macrophages play an important role in rendering resting T cells permissive for infection, in spreading HIV-1 to T cells, and in the pathogenesis of AIDS dementia. During highly active anti-retroviral treatment (HAART), macrophages keep producing virus because tissue penetration of antiretrovirals is suboptimal and the efficacy of some is reduced. Thus, to cure HIV-1 infection with antiretrovirals we will also need to efficiently inhibit viral replication in macrophages. The majority of the current drugs block the action of viral enzymes, whereas there is an abundance of yet unidentified host factors that could be targeted. We here present results from a genome-wide association study identifying novel genetic polymorphisms that affect in vitro HIV-1 replication in macrophages.
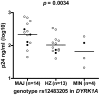
Methodology/principal findings: Monocyte-derived macrophages from 393 blood donors were infected with HIV-1 and viral replication was determined using Gag p24 antigen levels. Genomic DNA from individuals with macrophages that had relatively low (n = 96) or high (n = 96) p24 production was used for SNP genotyping with the Illumina 610 Quad beadchip. A total of 494,656 SNPs that passed quality control were tested for association with HIV-1 replication in macrophages, using linear regression. We found a strong association between in vitro HIV-1 replication in monocyte-derived macrophages and SNP rs12483205 in DYRK1A (p = 2.16 × 10(-5)). While the association was not genome-wide significant (p<1 × 10(-7)), we could replicate this association using monocyte-derived macrophages from an independent group of 31 individuals (p = 0.0034). Combined analysis of the initial and replication cohort increased the strength of the association (p = 4.84 × 10(-6)). In addition, we found this SNP to be associated with HIV-1 disease progression in vivo in two independent cohort studies (p = 0.035 and p = 0.0048).
Conclusions/significance: These findings suggest that the kinase DYRK1A is involved in the replication of HIV-1, in vitro in macrophages as well as in vivo.
Conflict of interest statement
Figures


Similar articles
-
The Dual-Specificity Kinase DYRK1A Modulates the Levels of Cyclin L2 To Control HIV Replication in Macrophages.J Virol. 2020 Feb 28;94(6):e01583-19. doi: 10.1128/JVI.01583-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31852782 Free PMC article.
-
Polymorphism in HIV-1 dependency factor PDE8A affects mRNA level and HIV-1 replication in primary macrophages.Virology. 2011 Nov 10;420(1):32-42. doi: 10.1016/j.virol.2011.08.013. Epub 2011 Sep 13. Virology. 2011. PMID: 21920574
-
ZNRD1 (zinc ribbon domain-containing 1) is a host cellular factor that influences HIV-1 replication and disease progression.Clin Infect Dis. 2010 Apr 1;50(7):1022-32. doi: 10.1086/651114. Clin Infect Dis. 2010. PMID: 20192730
-
Permissive factors for HIV-1 infection of macrophages.J Leukoc Biol. 2000 Sep;68(3):303-10. J Leukoc Biol. 2000. PMID: 10985244 Review.
-
Host hindrance to HIV-1 replication in monocytes and macrophages.Retrovirology. 2010 Apr 7;7:31. doi: 10.1186/1742-4690-7-31. Retrovirology. 2010. PMID: 20374633 Free PMC article. Review.
Cited by
-
A macrophage-cell model of HIV latency reveals the unusual importance of the bromodomain axis.Virol J. 2024 Apr 5;21(1):80. doi: 10.1186/s12985-024-02343-9. Virol J. 2024. PMID: 38581045 Free PMC article.
-
Truncation and Activation of Dual Specificity Tyrosine Phosphorylation-regulated Kinase 1A by Calpain I: A MOLECULAR MECHANISM LINKED TO TAU PATHOLOGY IN ALZHEIMER DISEASE.J Biol Chem. 2015 Jun 12;290(24):15219-37. doi: 10.1074/jbc.M115.645507. Epub 2015 Apr 27. J Biol Chem. 2015. PMID: 25918155 Free PMC article.
-
Immunogenetics of HIV disease.Immunol Rev. 2013 Jul;254(1):245-64. doi: 10.1111/imr.12071. Immunol Rev. 2013. PMID: 23772624 Free PMC article. Review.
-
Minding the gap in HIV host genetics: opportunities and challenges.Hum Genet. 2020 Jun;139(6-7):865-875. doi: 10.1007/s00439-020-02177-9. Epub 2020 May 14. Hum Genet. 2020. PMID: 32409920 Free PMC article. Review.
-
Dual-specificity tyrosine-phosphorylation regulated kinase 1A Gene Transcription is regulated by Myocyte Enhancer Factor 2D.Sci Rep. 2017 Aug 3;7(1):7240. doi: 10.1038/s41598-017-07655-1. Sci Rep. 2017. PMID: 28775333 Free PMC article.
References
-
- Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L, et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282:1627–1632. - PubMed
-
- Fischer M, Gunthard HF, Opravil M, Joos B, Huber W, et al. Residual HIV-RNA levels persist for up to 2.5 years in peripheral blood mononuclear cells of patients on potent antiretroviral therapy. AIDS Res Hum Retroviruses. 2000;16:1135–1140. - PubMed
-
- Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI035042/AI/NIAID NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- UO1-AI-35042/AI/NIAID NIH HHS/United States
- T32 AI007140/AI/NIAID NIH HHS/United States
- UO1-AI-35040/AI/NIAID NIH HHS/United States
- UO1-AI-35041/AI/NIAID NIH HHS/United States
- U01 AI035041/AI/NIAID NIH HHS/United States
- R37 AI047734/AI/NIAID NIH HHS/United States
- UL1-RR025005/RR/NCRR NIH HHS/United States
- UO1-AI-35039/AI/NIAID NIH HHS/United States
- U01 AI035040/AI/NIAID NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
- UO1-AI-35043/AI/NIAID NIH HHS/United States
- U01 AI035043/AI/NIAID NIH HHS/United States
- P30 AI027757/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

