Analysis of membrane topology and identification of essential residues for the yeast endoplasmic reticulum inositol acyltransferase Gwt1p
- PMID: 21367863
- PMCID: PMC3077662
- DOI: 10.1074/jbc.M110.193490
Analysis of membrane topology and identification of essential residues for the yeast endoplasmic reticulum inositol acyltransferase Gwt1p
Abstract
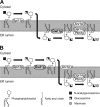
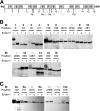
Glycosylphosphatidylinositol (GPI) is a post-translational modification that anchors cell surface proteins to the plasma membrane, and GPI modifications occur in all eukaryotes. Biosynthesis of GPI starts on the cytoplasmic face of the endoplasmic reticulum (ER) membrane, and GPI precursors flip from the cytoplasmic side to the luminal side of the ER, where biosynthesis of GPI precursors is completed. Gwt1p and PIG-W are inositol acyltransferases that transfer fatty acyl chains to the inositol moiety of GPI precursors in yeast and mammalian cells, respectively. To ascertain whether flipping across the ER membrane occurs before or after inositol acylation of GPI precursors, we identified essential residues of PIG-W and Gwt1p and determined the membrane topology of Gwt1p. Guided by algorithm-based predictions of membrane topology, we experimentally identified 13 transmembrane domains in Gwt1p. We found that Gwt1p, PIG-W, and their orthologs shared four conserved regions and that these four regions in Gwt1p faced the luminal side of the ER membrane. Moreover, essential residues of Gwt1p and PIG-W faced the ER lumen or were near the luminal edge of transmembrane domains. The membrane topology of Gwt1p suggested that inositol acylation occurred on the luminal side of the ER membrane. Rather than stimulate flipping of the GPI precursor across the ER membrane, inositol acylation of GPI precursors may anchor the precursors to the luminal side of the ER membrane, preventing flip-flops.
Figures




Similar articles
-
PIG-W is critical for inositol acylation but not for flipping of glycosylphosphatidylinositol-anchor.Mol Biol Cell. 2003 Oct;14(10):4285-95. doi: 10.1091/mbc.e03-03-0193. Epub 2003 Jun 13. Mol Biol Cell. 2003. PMID: 14517336 Free PMC article.
-
GWT1 gene is required for inositol acylation of glycosylphosphatidylinositol anchors in yeast.J Biol Chem. 2003 Jun 27;278(26):23639-47. doi: 10.1074/jbc.M301044200. Epub 2003 Apr 24. J Biol Chem. 2003. PMID: 12714589
-
MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast.Mol Biol Cell. 1999 Mar;10(3):627-48. doi: 10.1091/mbc.10.3.627. Mol Biol Cell. 1999. PMID: 10069808 Free PMC article.
-
Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids.J Lipid Res. 2007 May;48(5):993-1011. doi: 10.1194/jlr.R700002-JLR200. Epub 2007 Mar 14. J Lipid Res. 2007. PMID: 17361015 Review.
-
GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics.Biochim Biophys Acta. 2012 Aug;1821(8):1050-8. doi: 10.1016/j.bbalip.2012.01.004. Epub 2012 Jan 11. Biochim Biophys Acta. 2012. PMID: 22265715 Review.
Cited by
-
Chemical Genomics-Based Antifungal Drug Discovery: Targeting Glycosylphosphatidylinositol (GPI) Precursor Biosynthesis.ACS Infect Dis. 2015 Jan 9;1(1):59-72. doi: 10.1021/id5000212. Epub 2014 Dec 12. ACS Infect Dis. 2015. PMID: 26878058 Free PMC article.
-
Hedgehog Acyltransferase Promotes Uptake of Palmitoyl-CoA across the Endoplasmic Reticulum Membrane.Cell Rep. 2019 Dec 24;29(13):4608-4619.e4. doi: 10.1016/j.celrep.2019.11.110. Cell Rep. 2019. PMID: 31875564 Free PMC article.
-
Novel Promising Antifungal Target Proteins for Conquering Invasive Fungal Infections.Front Microbiol. 2022 Jun 16;13:911322. doi: 10.3389/fmicb.2022.911322. eCollection 2022. Front Microbiol. 2022. PMID: 35783432 Free PMC article. Review.
-
Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling.J Lipid Res. 2016 Jan;57(1):6-24. doi: 10.1194/jlr.R063313. Epub 2015 Nov 12. J Lipid Res. 2016. PMID: 26563290 Free PMC article. Review.
-
Chemical crosslinking and mass spectrometry to elucidate the topology of integral membrane proteins.PLoS One. 2017 Oct 26;12(10):e0186840. doi: 10.1371/journal.pone.0186840. eCollection 2017. PLoS One. 2017. PMID: 29073188 Free PMC article.
References
-
- Kinoshita T., Inoue N. (2000) Curr. Opin. Chem. Biol. 4, 632–638 - PubMed
-
- Pittet M., Conzelmann A. (2007) Biochim. Biophys. Acta 1771, 405–420 - PubMed
-
- Orlean P., Menon A. K. (2007) J. Lipid Res. 48, 993–1011 - PubMed
-
- Caro L. H., Tettelin H., Vossen J. H., Ram A. F., van den Ende H., Klis F. M. (1997) Yeast 13, 1477–1489 - PubMed
-
- Kinoshita T., Inoue N., Takeda J. (1995) Adv. Immunol. 60, 57–103 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

