MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors
- PMID: 21368052
- PMCID: PMC6623912
- DOI: 10.1523/JNEUROSCI.5085-10.2011
MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors
Abstract
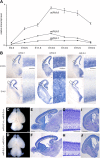
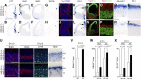
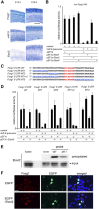
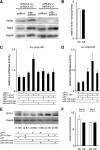
microRNA-9-2 and microRNA-9-3 double-mutant mice demonstrate that microRNA-9 (miR-9) controls neural progenitor proliferation and differentiation in the developing telencephalon by regulating the expression of multiple transcription factors. As suggested by our previous study, the Foxg1 expression was elevated, and the production of Cajal-Retzius cells and early-born neurons was suppressed in the miR-9-2/3 double-mutant pallium. At embryonic day 16.5 (E16.5), however, the Foxg1 expression was no longer elevated. The expression of an AU-rich RNA-binding protein Elavl2 increased at E16.5, Elav2 associated with Foxg1 3' untranslated region (UTR), and it countered the Foxg1 suppression by miR-9. Later, progenitor proliferation was reduced in the miR-9-2/3 double-mutant pallium with the decrease in Nr2e1 and Pax6 expression and the increase in Meis2 expression. The analyses suggest that microRNA-9 indirectly inhibits Pax6 expression by suppressing Meis2 expression. In contrast, together with Elavl1 and Msi1, microRNA-9 targets Nr2e1 mRNA 3' UTR to enhance the expression. Concomitantly, cortical layers were reduced, each cortical projection was malformed, and the tangential migration of interneurons into the pallium was impaired in the miR-9-2/3 double mutants. miR-9 also targets Gsh2 3' UTR, and Gsh2, as well as Foxg1, expression was elevated in the miR-9-2/3 double-mutant subpallium. The subpallium progenitor proliferation was enhanced, and the development of basal ganglia including striatum and globus pallidus was suppressed. Pallial/subpallial boundary shifted dorsally, and the ventral pallium was lost. Corridor was malformed, and thalamocortical and corticofugal axons were misrouted in the miR-9-2/3 double mutants.
Figures






References
-
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42.
-
- Agoston Z, Schulte D. Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development. 2009;136:3311–3322. - PubMed
-
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
