Bacterial community structures are unique and resilient in full-scale bioenergy systems
- PMID: 21368115
- PMCID: PMC3053989
- DOI: 10.1073/pnas.1015676108
Bacterial community structures are unique and resilient in full-scale bioenergy systems
Abstract
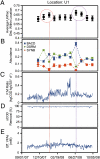
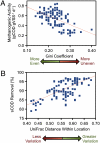
Anaerobic digestion is the most successful bioenergy technology worldwide with, at its core, undefined microbial communities that have poorly understood dynamics. Here, we investigated the relationships of bacterial community structure (>400,000 16S rRNA gene sequences for 112 samples) with function (i.e., bioreactor performance) and environment (i.e., operating conditions) in a yearlong monthly time series of nine full-scale bioreactor facilities treating brewery wastewater (>20,000 measurements). Each of the nine facilities had a unique community structure with an unprecedented level of stability. Using machine learning, we identified a small subset of operational taxonomic units (OTUs; 145 out of 4,962), which predicted the location of the facility of origin for almost every sample (96.4% accuracy). Of these 145 OTUs, syntrophic bacteria were systematically overrepresented, demonstrating that syntrophs rebounded following disturbances. This indicates that resilience, rather than dynamic competition, played an important role in maintaining the necessary syntrophic populations. In addition, we explained the observed phylogenetic differences between all samples on the basis of a subset of environmental gradients (using constrained ordination) and found stronger relationships between community structure and its function rather than its environment. These relationships were strongest for two performance variables--methanogenic activity and substrate removal efficiency--both of which were also affected by microbial ecology because these variables were correlated with community evenness (at any given time) and variability in phylogenetic structure (over time), respectively. Thus, we quantified relationships between community structure and function, which opens the door to engineer communities with superior functions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Angenent LT, Karim K, Al-Dahhan MH, Wrenn BA, Domíguez-Espinosa R. Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends Biotechnol. 2004;22:477–485. - PubMed
-
- Lettinga G. Anaerobic digestion and wastewater treatment systems. Antonie van Leeuwenhoek. 1995;67:3–28. - PubMed
-
- Leitão RC, van Haandel AC, Zeeman G, Lettinga G. The effects of operational and environmental variations on anaerobic wastewater treatment systems: A review. Bioresour Technol. 2006;97:1105–1118. - PubMed
-
- Briones A, Raskin L. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr Opin Biotechnol. 2003;14:270–276. - PubMed
-
- Rivière D, et al. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009;3:700–714. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

