Uncharacterized conserved motifs outside the HD-Zip domain in HD-Zip subfamily I transcription factors; a potential source of functional diversity
- PMID: 21371298
- PMCID: PMC3060862
- DOI: 10.1186/1471-2229-11-42
Uncharacterized conserved motifs outside the HD-Zip domain in HD-Zip subfamily I transcription factors; a potential source of functional diversity
Abstract
Background: Plant HD-Zip transcription factors are modular proteins in which a homeodomain is associated to a leucine zipper. Of the four subfamilies in which they are divided, the tested members from subfamily I bind in vitro the same pseudopalindromic sequence CAAT(A/T)ATTG and among them, several exhibit similar expression patterns. However, most experiments in which HD-Zip I proteins were over or ectopically expressed under the control of the constitutive promoter 35S CaMV resulted in transgenic plants with clearly different phenotypes. Aiming to elucidate the structural mechanisms underlying such observation and taking advantage of the increasing information in databases of sequences from diverse plant species, an in silico analysis was performed. In addition, some of the results were also experimentally supported.
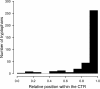
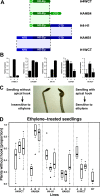
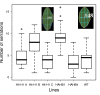
Results: A phylogenetic tree of 178 HD-Zip I proteins together with the sequence conservation presented outside the HD-Zip domains allowed the distinction of six groups of proteins. A motif-discovery approach enabled the recognition of an activation domain in the carboxy-terminal regions (CTRs) and some putative regulatory mechanisms acting in the amino-terminal regions (NTRs) and CTRs involving sumoylation and phosphorylation. A yeast one-hybrid experiment demonstrated that the activation activity of ATHB1, a member of one of the groups, is located in its CTR. Chimerical constructs were performed combining the HD-Zip domain of one member with the CTR of another and transgenic plants were obtained with these constructs. The phenotype of the chimerical transgenic plants was similar to the observed in transgenic plants bearing the CTR of the donor protein, revealing the importance of this module inside the whole protein.
Conclusions: The bioinformatical results and the experiments conducted in yeast and transgenic plants strongly suggest that the previously poorly analyzed NTRs and CTRs of HD-Zip I proteins play an important role in their function, hence potentially constituting a major source of functional diversity among members of this subfamily.
Figures





References
-
- Gong W, Shen YP, Ma LG, Pan Y, Du YL, Wang DH, Yang JY, Hu LD, Liu XF, Dong CX, Ma L, Chen YH, Yang XY, Gao Y, Zhu D, Tan X, Mu JY, Zhang DB, Liu YL, Dinesh-Kumar SP, Li Y, Wang XP, Gu HY, Qu LJ, Bai SN, Lu YT, Li JY, Zhao JD, Zuo J, Huang H, Deng XW, Zhu YX. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004;135:773–782. doi: 10.1104/pp.104.042176. - DOI - PMC - PubMed
-
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

