Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis
- PMID: 21371479
- PMCID: PMC3440003
- DOI: 10.1016/j.jmb.2011.01.018
Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis
Abstract
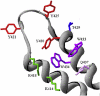
Initiation of RNA synthesis from DNA templates by RNA polymerase (RNAP) is a multi-step process, in which initial recognition of promoter DNA by RNAP triggers a series of conformational changes in both RNAP and promoter DNA. The bacterial RNAP functions as a molecular isomerization machine, using binding free energy to remodel the initial recognition complex, placing downstream duplex DNA in the active site cleft and then separating the nontemplate and template strands in the region surrounding the start site of RNA synthesis. In this initial unstable "open" complex the template strand appears correctly positioned in the active site. Subsequently, the nontemplate strand is repositioned and a clamp is assembled on duplex DNA downstream of the open region to form the highly stable open complex, RP(o). The transcription initiation factor, σ(70), plays critical roles in promoter recognition and RP(o) formation as well as in early steps of RNA synthesis.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

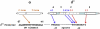
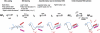
References
-
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. - PubMed
-
- Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, et al. Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol. Microbiol. 2008;68:538–546. - PubMed
-
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

