Endosomal targeting of the phosphoinositide 3-phosphatase MTMR2 is regulated by an N-terminal phosphorylation site
- PMID: 21372139
- PMCID: PMC3091194
- DOI: 10.1074/jbc.M110.209122
Endosomal targeting of the phosphoinositide 3-phosphatase MTMR2 is regulated by an N-terminal phosphorylation site
Abstract
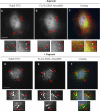
MTMR2 is a member of the myotubularin family of inositol lipid phosphatases, a large protein-tyrosine phosphatase subgroup that is conserved from yeast to humans. Furthermore, the peripheral neuromuscular disease Charcot-Marie Tooth disease type 4B has been attributed to mutations in the mtmr2 gene. Because the molecular mechanisms regulating MTMR2 have been poorly defined, we investigated whether reversible phosphorylation might regulate MTMR2 function. We used mass spectrometry-based methods to identify a high stoichiometry phosphorylation site on serine 58 of MTMR2. Phosphorylation at Ser(58), or a phosphomimetic S58E mutation, markedly decreased MTMR2 localization to endocytic vesicular structures. In contrast, a phosphorylation-deficient MTMR2 mutant (S58A) displayed constitutive localization to early endocytic structures. This localization pattern was accompanied by displacement of a PI(3)P-specific sensor protein and an increase in signal transduction pathways. Thus, MTMR2 phosphorylation is likely to be a critical mechanism by which MTMR2 access to its lipid substrate(s) is temporally and spatially regulated, thereby contributing to the control of downstream endosome maturation events.
Figures

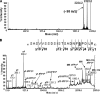






 , HEK293). The values are shown as fold-change from control ± S.D. (n = 3).
, HEK293). The values are shown as fold-change from control ± S.D. (n = 3).
References
-
- Barford D., Das A. K., Egloff M. P. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 133–164 - PubMed
-
- Fauman E. B., Saper M. A. (1996) Trends Biochem. Sci. 21, 413–417 - PubMed
-
- Guan K. L., Broyles S. S., Dixon J. E. (1991) Nature 350, 359–362 - PubMed
-
- Guan K. L., Dixon J. E. (1991) J. Biol. Chem. 266, 17026–17030 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

