Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms
- PMID: 21372847
- PMCID: PMC3178436
- DOI: 10.1038/cdd.2011.16
Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms
Abstract
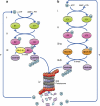
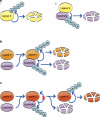
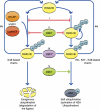
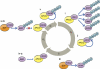
Ubiquitin modification of many cellular proteins targets them for proteasomal degradation, but in addition can also serve non-proteolytic functions. Over the last years, a significant progress has been made in our understanding of how modification of the substrates of the ubiquitin system is regulated. However, little is known on how the ubiquitin system that is comprised of ∼1500 components is regulated. Here, we discuss how the biggest subfamily within the system, that of the E3 ubiquitin ligases that endow the system with its high specificity towards the numerous substrates, is regulated and in particular via self-regulation mediated by ubiquitin modification. Ligases can be targeted for degradation in a self-catalyzed manner, or through modification mediated by an external ligase(s). In addition, non-proteolytic functions of self-ubiquitination, for example activation of the ligase, of E3s are discussed.
Figures
References
-
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. - PubMed
-
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-CBL/SLI-1. Mol Cell. 1999;4:1029–1040. - PubMed
-
- Vodermaier HC. APC/C and SCF: controlling each other and the cell cycle. Curr Biol. 2004;14:R787–R796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

